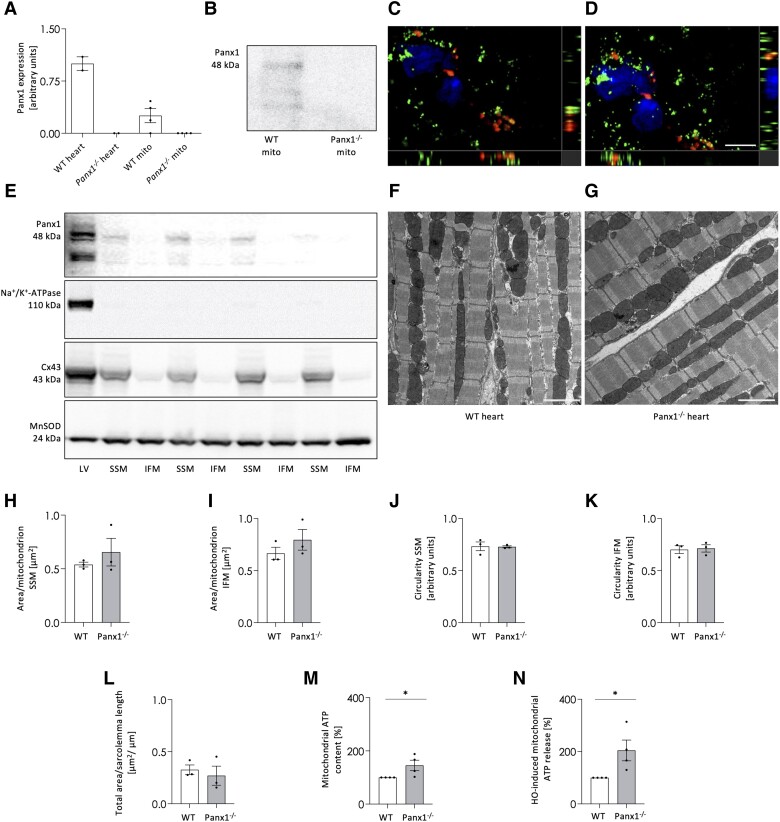
Figure 4.
Panx1 is present in cardiac SSM. qPCR shows Panx1 mRNA in mitochondria derived from WT ventricular tissue while absent in mitochondria of Panx1−/− ventricles. N = 4. mRNA extracted from WT and Panx1−/− hearts were used as positive and negative control, respectively (A). Panx1 protein is present in mitochondria isolated from WT ventricular tissue while absent in cardiomyocyte mitochondria of Panx1−/− ventricles (B). RNAscope in situ hybridization on murine heart cryosections demonstrated colocalization of Panx1 mRNA (red) with mitochondrial COX-I mRNA (green) (C–D). DAPI (blue). Scale bar = 5 μm. Co-localization is revealed by the yellow signal. (C and D) are representative photographs of three independent experiments. Western blots showing Panx1 protein in SSM fractions and not in IFM fractions isolated from ventricles of WT hearts (E). The absence of Na+/K + -ATPase confirmed the purity of mitochondrial extraction, the presence of Cx43 confirmed the correct subfractioning and MnSOD served as loading control. LV represents a total protein sample of LV tissue of the WT heart. Representative EM images of WT (F) and Panx1−/− (G) hearts. Scale bars = 2 µm. Morphological parameters, i.e. average area per SSM (H) or IFM (I) and circularity of SSM (J) or IFM (K), were not different between genotypes (WT: 44 images analysed from three hearts; Panx1−/−: 43 images analysed from three hearts). The total number of mitochondria analysed was 542 SSM and 668 IFM for WT hearts, and 356 SSM and 635 IFM for Panx1−/− hearts. The total SSM area relative to the length of the sarcolemma (L) was also not different between WT and Panx1−/− hearts. Statistical analysis of EM data was performed using a linear mixed model with either repeated measures and compound symmetry (mitochondrial area per sarcolemmal length) or a nested design (area per mitochondrion and shape circularity). The ATP content in Panx1−/− mitochondria was higher than in WT mitochondria. N = 4 (M). A hypo-osmotic shock induced more ATP release from Panx1−/− mitochondria than from WT mitochondria. N = 4, unpaired t-test (N).