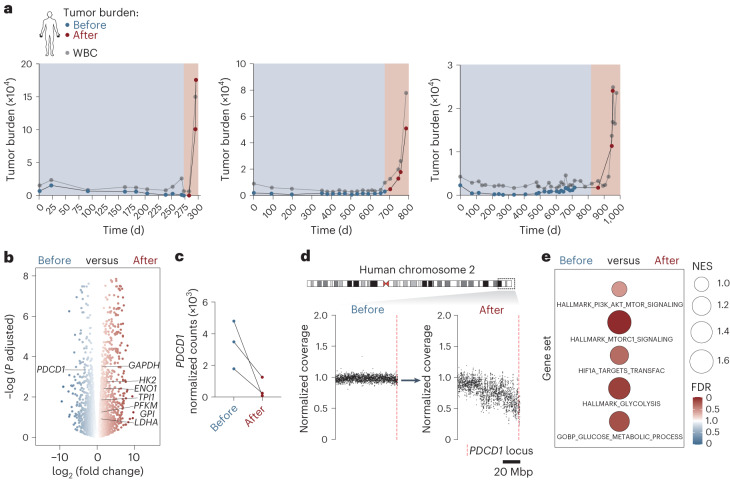
Fig. 6. PD-1 inactivation and glycolytic switch in hyperprogressive T-NHLs.
a, Serial longitudinal analysis of the tumor burden index and white blood cell (WBC) count for three individual patients with CTCL and hyperprogressive disease. Hyperprogression, rapid increase in tumor burden index (‘after’, shaded in red) after an extended period of stable disease course (‘before’, shaded in blue). b, Differential gene expression analysis of RNA-seq data derived from the three patients with CTCL in a before and after hyperprogression. Data points for selected glycolysis-related genes and PDCD1 are indicated. P, two-sided Wald test. c, RNA-seq read count of PDCD1 transcripts before and after hyperprogression shown for each patient individually. d, Whole-genome sequencing-based copy number aberration analysis of PDCD1-containing chromosomal region 2q36.3–2q37.3. The time point of genomic DNA isolation relative to hyperprogression is indicated (‘before’ versus ‘after’). The patient shown corresponds to the first patient in a. Top: the dashed box indicates the genomic region on human chromosome 2 that is shown in detail at the bottom. Bottom: copy number aberration analysis. The location of the PDCD1 locus is shown. e, GSEA for the indicated gene sets based on RNA-seq data derived from three patients with hyperprogression (a). The time point of RNA extraction relative to hyperprogression is indicated (‘before’ versus ‘after’). FDR, color intensity of circles; NES, circle diameter. Blue and red indicate the group in which a signature was positively enriched. a, Longitudinally collected data from three individual patients. b–e, Data from a single experiment with three patients.