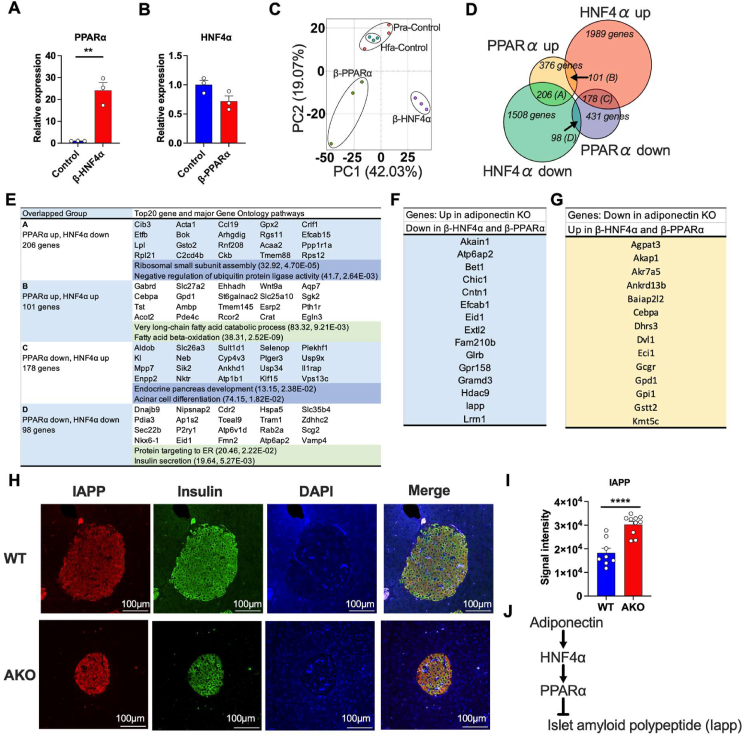
Figure 6.
Gene signatures of β-PPARα and β-HNF4α islets exhibit the potential pathways of adiponectin actions on pancreatic islets.(A) PPARα mRNA expression in β-HNF4α pancreatic islets. (n = 3) (B) HNF4α mRNA expression in β-PPARα pancreatic islets. (n = 3) (C) Principal component analysis of transcriptome signatures of β-PPARα and β-HNF4α pancreatic islets. (D) Overlap of the significantly up-regulated or down-regulated genes in β-PPARα and β-HNF4α pancreatic islets. (E) The major GO pathways and GO pathways highly affected by β-PPARα and β-HNF4α overexpression in each group. The numbers in the brackets describe fold-enrichment and FDR, respectively. (F) The list of genes that was significantly up-regulated in adiponectin KO islets but down-regulated in β-PPARα and β-HNF4α pancreatic islets. (G) The list of genes that was significantly down-regulated in adiponectin KO islets but up-regulated in β-PPARα and β-HNF4α pancreatic islets. (H) Representative image of immunofluorescence of the adiponectin KO pancreas. IAPP (red), Insulin (green) and DAPI (blue). Scale bar indicates 100 μm. (I) The calculation of the average signal intensity of IAPP immunofluorescence. (n = 8–10) (J) The schematic representation of potential adiponectin action cascade on the pancreatic islets through HNF4α and PPARα. Data are mean ± SEM. ∗∗P < 0.01 and ∗∗∗∗P < 0.0001. Unpaired 2-tailed t-tests were utilized for determining the p-values for (A) and (I).