Abstract

The 18 kDa translocator protein (TSPO) has gained considerable attention as a clinical biomarker for neuroinflammation and a potential therapeutic target. However, the mechanisms by which TSPO associates with ligands, particularly the endogenous porphyrin ligand protoporphyrin IX (PpIX), remain poorly understood. In this study, we employed mutagenesis- and spectroscopy-based functional assays to investigate TSPO-mediated photo-oxidative degradation of PpIX and identify key residues involved in the reaction. We provide structural evidence using electron spin resonance, which sheds light on the highly conserved intracellular loop (LP1) connecting transmembrane 1 (TM1) and TM2. Our findings show that LP1 does not act as a lid to regulate ligand binding; instead, it interacts strongly with the TM3–TM4 linker (LP3) to stabilize the local structure of LP3. This LP1–LP3 interaction is crucial for maintaining the binding pocket structure, which is essential for proper ligand binding. Our results also demonstrate that PpIX accesses the pocket through the lipid bilayer without requiring conformational changes in TSPO. This study provides an improved understanding of TSPO-mediated PpIX degradation, highlighting potential therapeutic strategies to regulate the reaction.
Keywords: tryptophan-rich sensory protein, porphyrin, spin label, photo-oxidation, glioblastoma
Introduction
The translocator protein (TSPO), also known as the peripheral benzodiazepine receptor (PBR) and tryptophan-rich sensory protein (TSPO), is an evolutionarily well-conserved protein with homologues present across organisms.1−5 TSPO is implicated in a variety of fundamental cellular processes, including the modulation of redox stress-related signaling, immune response, steroid synthesis, and apoptosis.4−7 TSPO is often found overexpressed in inflammatory and neurological diseases (e.g., Alzheimer’s and Parkinson’s diseases) as well as in glioblastoma, one of the most aggressive brain tumors.8−11 Thus, TSPO is not only a pharmacological target but also a biomarker of reactive gliosis and inflammation associated with a variety of brain injuries and tumors.12,13 There are numerous synthetic TSPO ligands being developed and explored as agonists and antagonists for the treatment of various diseases including anxiety, depression, and neurodegenerative disorders. PK11195 (isoquinoline carboxamide), for example, is a ligand for TSPO and is often used in positron emission tomography to visualize brain inflammation in patients with neuronal damage.2,3,5,12 Identifying the mechanisms of how ligands/drugs bind to TSPO and affect TSPO activity is urgently needed in the field.14
Structures of TSPO from Bacillus cereus (Bc), Rhodobacter sphaeroides (Rs), and mice have been determined by means of X-ray crystallography and NMR spectroscopy techniques.1−3 Those determined structures share the common feature of having five transmembrane (TM1-5) helical folds and a relatively long, highly conserved loop (residues 25–43 in the BcTSPO sequence; hereafter denoted by LP1) connecting TM1 and TM2, with minor differences among them. Even though the mammalian TSPOs share less than 30% identity in protein sequence with their bacterial homologues, the functions and structural conformations of TSPO are basically conserved. Mammalian TSPO can replace bacterial TSPO’s function as an oxygen sensor and be modified to bind an aquaporin in a manner similar to a plant TSPO.15,16 Several models of bacterial TSPO mimicking a human disease-associated polymorphism (rs6971) have revealed important functions of TSPO, which include the ability to bind and cleave porphyrins, such as protoporphyrin IX (PpIX), and the preferential engagement with steroids, providing a potential pathogenic mechanism of TSPO polymorphism linked to psychiatric diseases in humans.1,7,17 The binding of porphyrins is the most widely reported function of TSPO in all organisms. Homologues of TSPO are found in a variety of organisms where a protein that binds porphyrins has been implicated in the oxidative stress response.6 TSPO downregulation is reported to enhance PpIX accumulation, which is a toxic event, in several cell lines.1,18−21 Previous studies suggest that TSPO may catalyze degradation of PpIX with the consumption of reactive oxygen species, which may originate from photoexcitation of PpIX itself or from cellular processes.18 PpIX is a key intermediate in the heme biosynthesis pathway and an important photosensitizer used in photodynamic therapy for cancer treatment.22 TSPO has potential therapeutic and pharmacological value. However, the molecular intricacies governing the association of PpIX with TSPO, the formation of a bound complex, and the subsequent degradation of PpIX by TSPO have been largely unexplored.
Accordingly, the present study investigates the structural and binding properties of the bacterial homologue BcTSPO reconstituted in membrane scaffold protein (MSP)-based lipid nanodiscs (see Methods for details). We evaluated the catalytic degradation reaction of PpIX mediated by both wild-type (WT) BcTSPO and BcTSPO variants carrying mutations at key residues. Moreover, we perform the pulsed double electron–electron resonance (DEER)23−28 technique to measure the distances between selected spin-labeled pairs in BcTSPO embedded in nanodiscs. Hereafter, we refer to BcTSPO as TSPO for simplicity. We unravel the conformations of TSPO in nanodiscs, both in the presence and absence of PpIX (i.e., holo versus apo states). Furthermore, DEER provides new insights into the role of the highly conserved LP1 in TSPO activity. This study offers structural insights into the binding process of PpIX to TSPO and elucidates the molecular details concerning key residues that are instrumental in determining the photo-oxidative degradation of PpIX.
Results
To examine the TSPO-mediated activity in PpIX degradation, we recorded time-course fluorescence spectra of an equimolar mixture of PpIX and TSPO-loaded nanodiscs. A representative fluorescence analysis of WT TSPO activity toward PpIX is shown (WT:PpIX 1:1, Figure 1A). It shows clearly that the ground-state peak at 633 nm of the PpIX fluorescence decreases with time, demonstrating the degradation of PpIX by the WT-loaded nanodiscs. In the absence of TSPO (i.e., a mixture of PpIX and empty nanodiscs, denoted by the PpIX control in Figure 1A), the peak at 633 nm corresponding to PpIX is invariant during light exposure. The PpIX chemical structure and a collection of control experiments are provided in Supporting Information (Figure S1).
Figure 1.
Fluorescence assays to evaluate the activity of TSPO. (A) Upper panel: time-dependent fluorescence spectra of PpIX incubated with WT:TSPO-loaded nanodiscs 1:1. Lower panel: the corresponding relative fluorescence data at 633 nm. The PpIX control is a measurement on a mixture of PpIX and empty nanodiscs. (B) Crystal structure of TSPO (PDB code 4ryq) and the mutation sites (green) studied. LP1 is yellow colored. The inset shows the interacting residue complex (W31-L35-K98-L100) identified, in this study, as important for the LP1–LP3 stabilization. Sequences of various TSPO homologues are provided to show that the LP1 and LP3 regions are highly conserved, and the LP1 contains a signature short helix. Relative fluorescence data at 633 nm reveal the PpIX degradation activity of samples (C) W51F and W138F, (D) 34/102- and 34/151-related variants, and (E) 23/35 and 23/151 TSPO. Note that the fluorescence data indicate that W51F and W138F degrade PpIX into different products, as detailed in Figure S1C. Except for the PpIX control, all measurements were performed with an equimolar mixture of PpIX and TSPO.
Identification of Key Residues in TSPO Embedded in the Nanodisc Lipid Bilayer
This study performed fluorescence analysis to examine the PpIX degradation activity of various TSPO mutants (Figure 1B). First, we conducted the measurements on the W-to-F mutation of a conserved tryptophan for respective W51F and W138F (both of which are conserved across all species). They were previously identified, in a fluorescence assay with detergent-solubilized BcTSPO, to be key tryptophan residues in association with ligands (PK11195 and PpIX),2 but they have not been measured in a nanodisc lipid bilayer. By incubating TSPO-loaded nanodiscs with PpIX, we found that TSPO-mediated PpIX degradation is clearly hampered by the W51F mutation and is largely abolished with the other mutation, W138F (Figures 1C and Figure S1C). Our nanodisc-based results demonstrate that W51 and W138 are important residues in the TSPO-mediated PpIX degradation and, moreover, reveal that the reaction is not basically altered between the surrounding environments (nanodisc lipids versus DDM detergent) (Figure S2A,B). This study also determined steady-state kinetic parameters for the PpIX degradation activity of WT TSPO (Figure S2C,D).
Role of the Highly Conserved LP1 in PpIX Degradation Activity of TSPO
There are several highly conserved residues in the LP1 region (Figure 1B), but the importance of LP1 in determining PpIX degradation has remained unclear. Next, we performed fluorescence analysis on several cysteine variants of TSPO. The reason that we prepared cysteine variants of TSPO is because this study, as given in a later section, performed cysteine mutagenesis and site-directed spin labeling to explore the structure–function relationship. All cysteine variants of TSPO were derived from the cysteine-free mutant (i.e., C107S), as the WT has only one native cysteine residue. Notably, we found that two double-cysteine variants, 34/102 and 34/151 (Figure 1D), retain the same catalytic activity as WT, but the activity is hampered if one additional mutation is introduced to impede the interaction of LP1 with other residues. With the mutation D33P, which was engineered to hamper the helicity of the short helix (residues 30–35; Figure 1B) on LP1 and hinder the local interactions, we observed a marked decrease in the activity of 34/102 (i.e., 34/102-D33P) to degrade PpIX (Figure 1D). The D33P is effective in disrupting the local environment of LP1. With the K98E mutation, which was engineered to invert the charge, we observed a similar marked reduction in the catalytic activity of 34/102 (i.e., 34/102-K98E in Figure 1D). These experimental results reveal the importance of the local interactions between the LP1 and the TM3-TM4 linker (hereafter denoted by LP3; inset in Figure 1B). In addition, structure-based analysis using the Arpeggio program29 indicates that there are various interacting residue pairs (e.g., W31-L35-K98-L100, shown in the inset of Figure 1B) between LP1 and LP3 in the crystal structure of TSPO, providing support for our experimental results with the D33P and K98E mutations. A detailed discussion of the Arpeggio analysis result is provided (Figure S3).
To further validate the finding presented above, we conducted a fluorescence assay on another double-cysteine variant, TSPO 23/35 (Figure 1E). Residue L35 is highly conserved in the LP1 sequence (as shown in Figure 1B) and is identified as a key residue in the W31-L35-K98-L100 complex determining the LP1–LP3 interaction. Consequently, its mutation would be expected to impede the LP1–LP3 interaction. We observed that the activity of 23/35 is significantly hampered, while the activity of 23/151, in which site 151 is known (as shown in Figure 1D) to cause an insignificant change in activity, remains largely unchanged compared to the WT. This indicates that TSPO 23/35 is functionally affected by the L35C mutation, which disrupts the W31-L35-K98-L100 complex and can thus be considered a loss-of-function cysteine variant. These results clearly demonstrate that the LP1–LP3 interaction, stabilized by the W31-L35-K98-L100 complex, plays a role in the TSPO-mediated PpIX degradation.
Roles of Key Residues in Ligand Binding and Degradation
Next, we explored the molecular details of how the LP1–LP3 interaction modulates the reaction. The recorded time-dependent fluorescence data were further analyzed to determine the respective reaction velocity (V0; see Figure 2A), as detailed in Methods. We observed that TSPO 34/102, 34/151, and 23/151 have a V0 value similar to that of WT (approximately 17 nM/s), which is much greater than those (approximately 5 nM/s) for W51F, 23/35, 34/102-D33P, 34/102-K98E, 34/151-D33P, and 34/151-K98E. The V0 value of TSPO toward PpIX was reduced further (to below 2 nM/s) if W138F-related variants (W138F and 34/151-W138F) were used. Clearly, a significant reduction in the reaction velocity V0 is observed only for the W138F-related variants.
Figure 2.
Reaction velocity and dissociation constant of TSPO toward PpIX. (A) Reaction velocity V0 of the PpIX degradation activity of TSPO variants. (B) Representative results of the tryptophan fluorescence quenching measurements. Solid lines are fits. (C) Dissociation constant Kd of the TSPO-PpIX complex determined in the tryptophan fluorescence quenching assay. Data of V0 and Kd are shown as the mean ± standard error (SE).
Moreover, we performed a tryptophan fluorescence quenching binding assay (Figure 2B; details given in the Methods) to obtain the dissociation constant (Kd) of PpIX with TSPO variants. We observed that some samples with a low V0 (e.g., W51F, 34/151-D33P, 34/151-K98E, 34/102-D33P, 34/102-K98E, and 23/35) have Kd values much higher (i.e., lower affinity for PpIX) than those of WT (Figure 2C). It indicates that the mutations of W51F, D33P, K98E, and L35C impede the formation of the TSPO-PpIX bound form and consequently have a reduced affinity for PpIX, leading to a slower reaction velocity. Regarding the result of D33P, K98E, and L35C, it also supports the idea that retaining the LP1–LP3 interaction is the key to promoting the formation of the PpIX-TSPO complex. Generally, the higher the Kd is, the slower the reaction is. Exceptions were observed for the W138F-related variants in which they were low in both V0 and Kd. It indicates that although W138F and 34/151-W138F bind with PpIX as strongly as WT, the degradation of the bound PpIX is hampered. It demonstrates that W138 plays a role in the catalytic degradation of PpIX rather than in the binding of PpIX to TSPO. Overall, our findings identify the different roles of W51 and W138 in TSPO activity and reveal a previously undescribed role for LP1. Specifically, we show that the stabilization between LP1 and LP3 is important in determining the binding of PpIX to TSPO.
Structural Differences between Apo and Holo TSPO
To explore possible structural changes between apo and holo states of TSPO, we performed DEER measurements on various double-labeled TSPO (Figure 3A) embedded in lipid nanodiscs, one at a time, and then analyzed the experimental DEER data (see Figure S4A for raw DEER traces) to obtain interspin distance distributions. The double-cysteine variants of TSPO studied have a similar PpIX degradation activity to WT (Figure S4B–D), suggesting that these cysteine variants are functionally active. In a comparison of the DEER distance distributions of apo and holo TSPO (blue and black lines in Figure 3B), we found mostly minor modifications of the distance distributions, suggesting that large conformational changes are not required to bind PpIX ligands. Those differences between apo and holo states are insignificant and within error bounds (Figure S5A,B).
Figure 3.
Spin-labeling sites and distance distributions. (A) Topology cartoon of TSPO. Green circles indicate the sites mutated to cysteines to prepare double-cysteine variants for spin-labeling study. (B) Distance distributions of double-labeled TSPO variants in nanodiscs, studied in apo and hole states of TSPO. Gray histograms are distance distributions predicted from the crystal (PDB code 4ryq).
Next, we explored the conformational differences between the TSPO in nanodiscs and the BcTSPO crystal (PDB ID 4ryq) grown from detergent-solubilized TSPO. Interspin distance distributions corresponding to the crystal structure were simulated (as illustrated by the gray histograms in Figure 3B) using the MtsslWizard computational tool.30 This program generated potential conformations of the spin label that do not interfere with the static crystal model of TSPO. While there are some minor differences in the distance distributions between the DEER and crystal studies, these discrepancies are too small to be modeled based on the DEER distance distributions due to the absence of suitable software. Crucially, these minor differences are considered insignificant, as TSPO’s PpIX degradation activity remains largely unchanged between the nanodisc and detergent environments (Figures S2 and S4B,C).
Structural Insights into the LP1-Mediated TSPO Activity
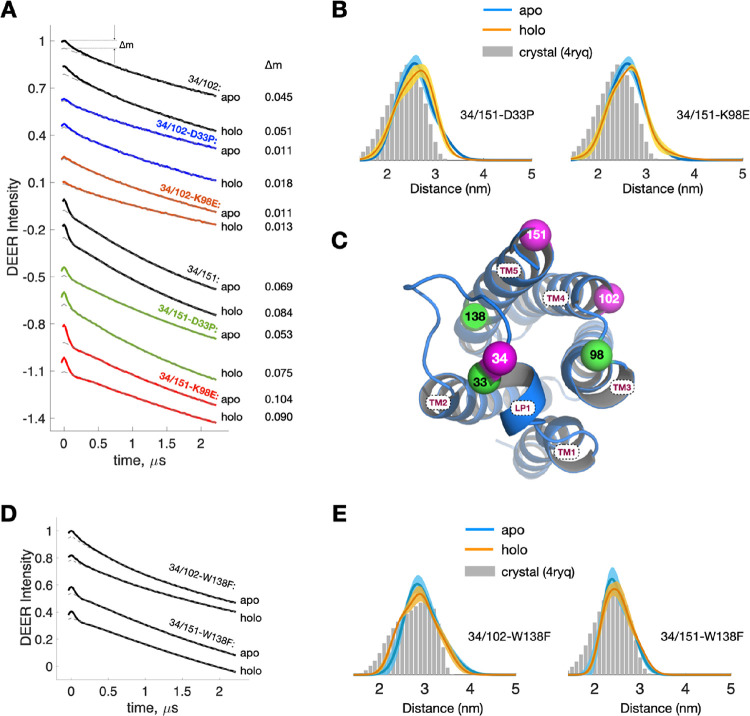
Based on the results of functional studies of TSPO (Figures 1 and 2), we performed DEER measurements on TSPO variants related to D33P and K98E to explore how the two mutations impede TSPO-mediated PpIX degradation. The most noteworthy aspect of the results is that the DEER traces of 34/102-D33P and 34/102-K98E suffer from a lack of clear dipolar modulation depths (Δm), which represents a substantial reduction in the modulation depth of the DEER trace (Figure 4A). The Δm of apo 34/102 is 0.045, and it is reduced by more than 70% with either the D33P or K98E mutation. The reduction in Δm suggests an increase in the intermolecular distances in 34/102. Figure S6A shows a control experiment verifying that the Δm reduction observed is not due to a D33P- or K98E-induced change in the labeling efficiency among cysteine variants. For these experimental DEER traces lacking distinct modulation depths, the standard method for DEER data analysis fails, and the extraction of precise distance distributions is precluded. This result provides support for the conclusion drawn from our fluorescence data and reveals a new finding on the 34/102. The single mutation, D33P or K98E, is effective in disrupting the LP1–LP3 stabilization, leading to an extended distance in 34/102.
Figure 4.
DEER measurements on TSPO variants with D33P, K98E, and W138F mutations. (A) Raw DEER traces of the 34-related studies. Modulation depths (Δm) are indicated. (B) Distance distributions of the double-labeled apo and holo TSPO variants, 34/151-D33P and 34/151-K98E. They are barely affected by the single mutation (D33P or K98E) and closely resemble the predictions based on the WT crystal. (C) Cartoon model showing the positions of spin-labeling sites (magenta) and the mutations (green) D33P, K98E, and W138F. (D) Raw DEER traces of 34/102 and 34/151 with the W138F mutation. (E) Distance distributions of 34/102-W138F and 34/151-W138F. Minor differences in the distance distributions of apo and holo TSPO are observed. The DEER results closely resemble the predictions based on the WT crystal.
Nevertheless, the modulation depths of 34/151-related DEER traces remain distinct and well-defined (Figure 4A) with either D33P or K98E. The distance distributions of 34/151-D33P and -K98E appear to differ slightly between the apo and holo states of TSPO (Figure 4B) and are virtually unchanged as compared to those (Figure 3B) obtained without the mutation. Also, they are similar to the distance distributions predicted from the crystal (Figure 4B). It indicates that the distances in 34/151 and respective local structures of the 34 and 151 sites are barely affected by either D33P or K98E. Together with the 34/102 results, it shows that the local environment of 102 is the most affected (Figure 4C), among the three sites (34, 102, and 151), by either of the mutations (D33P and K98E).
Role of W138 in the Structure of TSPO
As demonstrated by the activity and binding assays (Figure 2), we determined that W138 is involved in the catalytic degradation of PpIX rather than in the binding of PpIX to TSPO. To further elucidate the role of W138 in the structure of TSPO, we performed DEER measurements on the W138F variants (Figure 4D). The obtained DEER results for 34/102 and 34/151 with the W138F mutation closely resemble the distance distributions predicted from the crystal structure (Figure 4E). This suggests that the distances in 34/102 and 34/151 remain unaffected by the W138F mutation, implying that W138F has a minimal impact on the structural conformation of TSPO. Collectively, our findings support the notion that W138 primarily functions in catalyzing the degradation of PpIX and plays an insignificant role in maintaining structural stability.
Binding Cavity of TSPO for PpIX
By utilizing the protein cavity detection and evaluation program CavityPlus,31 we predicted that the central cavity is the most probable binding site (with the highest values of CavityScore, DrugScore, and Druggability) for PpIX (Figure 5A) and that the opening between TM1 and TM2 is the only pathway for ligand access into the cavity. The proposed cavity for PpIX is consistent with a recent crystal structure of a heme-binding RsTSPO (Figure S6B).32 Using the APBS (Adaptive Poisson–Boltzmann Solver) modeling program,33 we generated and visualized the electrostatic surface potentials of the cavity (Figure 5B). As seen, both of the predicted cavities overlap well and are suitable for binding PpIX. These observations suggest that the access of the PpIX ligand to the cavity occurs from the lipid bilayer.
Figure 5.
Predictions of the binding cavity and the verification. (A) Central cavity (green) of TSPO predicted with the CavityPlus program. The cavity can be accessed via the opening between the cavities of TM1 and TM2. (B) PpIX is modeled into the APBS-modeled cavity, shown in a transparent mesh. An isosurface mesh colored according to charges is drawn at 5.0 (blue) and −5.0 (red) kT/e electrostatic potentials, as highlighted in the inset. W51 and W138 are closely associated with PpIX. The prediction is consistent with the cavity in the heme-binding RsTSPO (Figure S6B). (C) PpIX degradation activity of TSPO at varying lipid fluidity. We measured the PpIX degradation activity of WT TSPO in various nanodiscs composed of different lipid compositions (as indicated). (D) Positions of sites S22 and A34 in TSPO. (E) Results of the activity assay for DDM-solubilized TSPO with and without PEG conjugation. (F) Results of the binding assay for DDM-solubilized TSPO with and without PEG conjugation. Data are shown as the mean ± standard error. Full fluorescence spectra are given in Figure S7.
To validate these predictions, we reduced the fluidity of the lipid bilayer by changing the state of the lipid phase from liquid-disordered (Ld) to gel states. At 300 K, an 80:20 molar ratio of DPPC:DLPC is in a gel state, while the other lipid mixtures studied are in an Ld state (Figure 5C). The TSPO activity remains relatively unchanged with different lipid compositions and types as long as the bilayer is in an Ld phase. However, the TSPO activity is substantially reduced in the gel phase, which is characterized by slower fluidity. These results highlight the importance of lipid fluidity in determining TSPO’s PpIX degradation activity and suggest that PpIX enters the cavity through the opening between TM1 and TM2 in the lipid bilayer.
To confirm the critical role of the opening between TM1 and TM2 for PpIX entry, we engineered two single-cysteine TSPO variants (S22C and A34C; see Figure 5D) and performed activity and binding assays on these variants, both with and without poly(ethylene glycol) (PEG, 5 kDa) conjugation, to block PpIX entry. Site 22 is located at the opening, while site 34 faces the solvent side on LP1. Our results demonstrate that the PpIX degradation activities of S22C, A34C, and A34C-PEG are comparable to those of WT, whereas the S22C-PEG activity is significantly hindered (Figure 5E). Conjugating PEG at site S22 effectively blocks PpIX entry into the cavity. The binding assay results confirm the blocking effect of PEG at site 22, as S22C-PEG has a Kd value approximately 10-fold higher than that of WT (Figure 5F). Our findings strongly support the view that PpIX enters the cavity through the opening between TM1 and TM2.
Discussion
LP1 Helps to Stabilize the Local Structure of LP3
Taken together, we hypothesize a mechanism whereby a single mutation, either D33P or K98E, impedes the binding of PpIX to TSPO. Our study demonstrates that LP1 of TSPO is a critical player in stabilizing the local structure of LP3 connecting TM3 and TM4. Specifically, the D33P or K98E mutation disrupts the LP1–LP3 interaction, primarily the W31-L35-K98-L100 complex, leading to a notable alteration in the local structure of the C-terminal region of LP3, where site 102 is located, and a consequent conformational change toward an extended structure. Notably, the disruption of the LP1–LP3 interaction has little effect on the local structure and the position of LP1. Our findings are supported by the analysis of interatomic interactions (Figure S3), which indicates that the C-terminal region of LP3 has substantially fewer interactions with LP1 than the N-terminal region of LP3. The LP1–LP3 stabilization is crucial for maintaining the structural integrity of LP3 and thus for retaining the binding pocket of TSPO. Given that LP1 is conserved across various species, our finding suggests a new direction for TSPO research: targeting this region with small molecules or peptides could modulate the TSPO activity and potentially have therapeutic effects.
PpIX Diffuses into the TSPO Cavity from the Lipid Bilayer
The absence of a reported structure of the TSPO-PpIX bound form specifically for BcTSPO leaves a gap in the knowledge of the binding pathway. However, this study sheds new light on the topic. Previous crystal structures of BcTSPO and RsTSPO indicated interacting pairs consisting of residues from LP1 and its nearby structures, such as W30 with M97, W30 with K36, D32 with R43, and W39 with G141 in RsTSPO.1,2 Therefore, LP1 was previously proposed to act as a lid of the central cavity of TSPO to regulate PpIX binding.3,4 Nevertheless, the findings presented here do not provide support for such an assertion. Our DEER data reveal that LP1’s position and structure remain largely unchanged before and after PpIX binding. Additionally, the experiments with D33P- and K98E-related variants show that disrupting LP1–LP3 stabilization significantly reduces TSPO’s binding affinity for PpIX. Thus, the idea of LP1 undergoing opening and closing is not supported by our data. These results further discount the notion that LP1 functioning as a lid is crucial for capturing free PpIX and facilitating the formation of the TSPO-PpIX bound form. Thus, PpIX access to TSPO’s central cavity must not occur through an LP1-gated pathway. Our fluorescence data demonstrate that the activity of TSPO in PpIX degradation can be effectively reduced by reducing the fluidity of the lipid bilayer or by blocking the proposed opening with PEG, suggesting that PpIX diffuses into the cavity through the opening between TM1 and TM2 in the lipid bilayer. This information is potentially useful in the design of drugs that either facilitate or inhibit binding of PpIX to TSPO, depending on the desired therapeutic outcome.
The TSPO Monomer Is Functionally Significant
TSPO has been reported to exist in both dimeric and monomeric states in humans, mice, and bacteria (such as BcTSPO and RsTSPO).1−3 The dimer interface varies between species, indicating that it is not conserved. However, an important question remains unanswered: is the dimer state functionally significant? Our DEER results reveal that BcTSPO remains as a monomer in the nanodisc lipid bilayer (Figure S8A). Moreover, the biochemical assays conducted in this study demonstrate that the monomeric state of TSPO is functionally significant. This finding suggests that targeting the monomeric form of TSPO could be a viable therapeutic strategy.
In summary, we investigated the mechanisms underlying the binding and degradation of PpIX by TSPO using various techniques, including mutagenesis, fluorescence, and DEER. Our results contradict the previous notion, suggested by static NMR and crystal structures, that LP1 operates as a lid to regulate PpIX binding. Rather, we discovered that LP1 is closely associated with LP3 during the reaction, which is crucial for maintaining the structural integrity of the binding cavity and the binding affinity of PpIX. Additionally, we identified specific residues, such as W138 and W51, that play a critical role in the photo-oxidation and binding/degradation of PpIX, respectively. Our research sheds light on the potential therapeutic significance of LP1, as it is conserved across various species. Our findings regarding the PpIX entry pathway and the critical role of LP1 can inform the development of novel therapeutics targeting TSPO.
Materials and Methods
Cloning and Site-Directed Mutagenesis
The gene encoding TSPO from Bacillus cereus was cloned into a pMCSG7 expression vector, which produces a 10-His tag at the N-terminus of BcTSPO. The DNA plasmid was a kind gift from Dr. Wayne A. Hendrickson (Columbia University). The cysteine-free construct of BcTSPO (C107S) was used to prepare various single- or double-cysteine variants, as well as other mutations such as W51F and W138F. Point mutations were generated using a QuikChange site-directed mutagenesis kit (Stratagene) with complementary oligonucleotide primers. All substitutions were verified by DNA sequencing. These plasmids were transformed into the Escherichia coli strain BL21 (DE3) pLysS competent cells (Agilent), which were used for protein expression.
Expression and Purification of BcTSPO
BcTSPO was expressed and purified as previously described with minor modifications.2 For large-scale expression and purification, we typically inoculated 25 mL of the starting culture in terrific broth (TB) media containing 50 μg/mL ampicillin and 25 μg/mL chloramphenicol with a glycerol stock of the BcTSPO-expressing bacteria. The starting culture was incubated at 37 °C with shaking. A dense overnight culture was subsequently used to inoculate 0.8 L of the antibiotic-containing TB media at a 1:200 dilution. Cultures were grown at 37 °C with shaking until the optical density at 600 nm (OD600) reached 0.6–0.8 and then induced with 1 mM isopropyl β-d-thiogalactopyranoside (IPTG). After induction with IPTG, the cultures were incubated at 20 °C for 18 h. Cells were harvested by centrifugation at 9000g for 5 min at 4 °C and stored at −80 °C.
The frozen cell pellets were resuspended in buffer A (50 mM HEPES, 300 mM NaCl, 20 mM imidazole, 1 mM MgCl2, and 5% glycerol, pH 7.8) with an additional 10 μg/mL DNaseI. The resuspended pellets were sonicated by pulse sonication at 70% output on ice for 15 min. The lysed cell solution was centrifuged at 12,800g for 50 min, and then, the supernatant was centrifuged at 45,000g for 1 h to collect membrane pellets. The membrane pellets were frozen at −20 °C or immediately homogenized in buffer A with an additional 1.5% (w/v) n-dodecyl-β-d-maltopyranoside (β-DDM). The resuspended membrane pellets were stirred gently at 4 °C for 1 h followed by centrifugation at 45,000g for 30 min to remove insoluble components. The cleared supernatant was filtrated through a 0.45 μm filter and passed through a 5 mL HisTrap HP nickel-affinity column (GE Healthcare) pre-equilibrated with buffer A with an additional 0.5% (w/v) β-DDM. The nickel-affinity column was washed with 20 column volumes (CVs) of buffer B (50 mM HEPES, 500 mM NaCl, 75 mM imidazole, 0.05% (w/v) β-DDM, and 5% glycerol, pH 8) and then eluted by 6 CVs of buffer C (50 mM HEPES, 300 mM NaCl, 500 mM imidazole, 0.05% (w/v) β-DDM, and 5% glycerol, pH 8). Fractions containing BcTSPO were collected and buffer-exchanged into buffer D (50 mM HEPES, 150 mM NaCl, and 0.05% (w/v) β-DDM, pH 8) to remove imidazole. Purified BcTSPO was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with Coomassie blue staining, and the protein concentration was determined using absorption spectroscopy at 280 nm.
Spin Labeling of ESR Samples
Following purification of BcTSPO, spin labeling was performed by the addition of a 25-fold molar excess of (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl) methanethiosulfonate (MTSSL) (Enzo Life Sciences) for each cysteine from a 0.1 M stock solution in acetonitrile. The labeling reaction was allowed to proceed in the dark at 4 °C overnight to yield a spin-label side chain R1. Unreacted spin labels were removed completely during the process of nanodisc sample preparation described later.
Expression and Purification of Membrane Scaffold Protein (MSP)
Membrane scaffold protein (MSP1D1, herein denoted by MSP for simplification) was expressed and purified as previously described with minor modifications.23,34,35 Briefly, the gene of MSP was cloned into a pET-28a (Addgene) expression vector, which produced a 7-His tag at the N-terminus of MSP. The plasmid was transformed into the Escherichia coli strain BL21 (DE3) pLysS competent cell (Agilent) for protein expression. A single colony from a fresh LB-agar plate was used to inoculate 20 mL of terrific broth (TB) media containing 30 μg/mL kanamycin, and the culture was incubated at 37 °C with shaking. After 4–6 h, OD600 approximately reached 0.6–0.8, and the starting culture was used immediately or stored at 4 °C for up to 18 h. Then, the starting culture was used to inoculate 0.8 L of an antibiotic-containing TB medium (30 μg/mL kanamycin). The culture was incubated at 37 °C with shaking for 4–5 h. When OD600 reached 1.5–2, induction with 1 mM IPTG was performed. After 1 h, the temperature was switched to 28 °C. After 2.5–3 h, the cells were collected by centrifugation and stored at −80 °C until further purification.
The cell pellets were resuspended in 30 mL of 20 mM sodium phosphate, 100 mM NaCl, 10 mM MgSO4, and 1% Triton X-100, pH 7.4, with an additional 10 μg/mL dNaseI and 300 μL of phenylmethylsulfonyl fluoride (PMSF) in 0.1 M ethanol stock, and then sonicated by pulse sonication at 70% output on ice for 15 min. Cell debris was removed by centrifugation at 30,000g for 30 min. The cleared supernatant was filtrated through a 0.45 μm filter and passed through a 5 mL HisTrap HP nickel-affinity column (GE Healthcare) pre-equilibrated with 5 CVs of 20 mM sodium phosphate, 100 mM NaCl, and 1% Triton X-100, pH 7.4. The nickel-affinity column was washed with the following three different buffers: (i) 6 CVs of 40 mM Tris-HCl, 300 mM NaCl, and 1% Triton X-100, pH 8.0; (ii) 6 CVs of 40 mM Tris-HCl, 300 mM NaCl, and 50 mM sodium cholate, pH 8.0; (iii) 6 CVs of 40 mM Tris-HCl, 300 mM NaCl, and 40 mM imidazole, pH 8.0. The bound MSP was eluted with 6 CVs of 40 mM Tris-HCl, 300 mM NaCl, and 400 mM imidazole, pH 8.0. Purified MSP was buffer-exchanged into MSP storage buffer (20 mM Tris-HCl and 100 mM NaCl, pH7.4) and concentrated to around 10 mg/mL using Amicon Ultra-10K centrifugal filter units. Protein purity was checked by SDS-PAGE with Coomassie blue staining, and the protein concentration was determined using absorption spectroscopy at 280 nm.
Reconstitution of BcTSPO into Nanodiscs
Lipids POPC (1-palmitoyl-2-oleoyl-glycero-3-phosphocholine) and POPE (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine) dissolved in chloroform were prepared in a 60:40 molar ratio, dried under nitrogen flow, and desiccated under vacuum for 6 h. Reconstitution of BcTSPO into nanodiscs was performed using a modified Sligar’s protocol.23,36,37 Purified BcTSPO in β-DDM micelles (buffer D) was mixed with appropriate lipids, MSP, and sodium cholate (SC) in the following molar ratio: TSPO/MSP/lipid/SC = 1/4/270/540. The mixture was incubated on ice for 30 min. To remove detergent, 1 g/mL Biobeads SM-2 (Bio-Rad) was added into the mixture and incubated at 4 °C overnight. Biobeads were removed by a short spin-down, and the nanodisc suspension was then run on a Superdex 200 Increase 10/300 GL gel filtration column (Cytiva) pre-equilibrated with buffer E (50 mM HEPES and 150 mM NaCl, pH 8.0). The BcTSPO-containing nanodiscs were collected and concentrated with Amicon Ultra-50K centrifugal filter units and characterized using SDS-PAGE to verify reconstitution (Figure S8B). In addition, we changed the lipid fluidity at 300 K by varying lipid compositions and types (Figure 5C) of the following: DPPC (1,2-dipalmitoyl-sn-glycero-3-phosphocholine), DLPC (1,2-dilauroyl-sn-glycero-3-phosphocholine), POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol), POPE, and POPC. The lipid compositions studied (Figure 5C) were as follows: POPC:POPE = 60:40, POPG:POPE = 80:20, 60:40, 40:60, and 20:80, and DPPC:DLPC = 80:20 and 20:80, where only the mixture DPPC:DLPC 80:20 is in a gel state and the rest are in a liquid-disordered state.38 The lipid reconstitution protocols for nanodiscs with high Tm lipids are optimized as previously described.35
Assay to Measure the PpIX Degradation Activity of TSPO (by Absorbance and Fluorescence)
Activity assays were performed as previously described.2,18 Protoporphyrin IX (PpIX) has a prominent Soret band feature peaked at 410 nm and weaker Q-band features with four peaks at 628, 574, 537, and 503 nm. We monitored the decay of the Soret band and Q bands to determine the PpIX degradation activity of BcTSPO. UV–vis scans of the solution mixtures of BcTSPO and PpIX showed two new peaks: one at 344 nm and another at 570 nm. A U-3900 spectrophotometer (Hitachi) was used to perform all the absorption spectra measurements.
PpIX displayed a characteristic fluorescence emission at 633 nm when excited at 405 nm. We used an F-7000 FL spectrophotometer (Hitachi) to perform all of the fluorescence spectra measurements. Experiments with this instrument were conducted as a series of pulses, where each comprised 60 s continuous illumination during recording. The reaction solution in a 1 mm square quartz cuvette contained PpIX and BcTSPO-loaded nanodiscs in a molar ratio of 1:1 with a total volume of 300 μL (Figure S9). For the controls, the sample buffer replaced the protein solution. The first three acquired data points (approximately 120 s duration) at 633 nm were fitted to a straight line. The absolute value of the slope is the initial reaction rate (denoted by V0). The experiments were performed in triplicate. Shown are averages with the standard error (SE).
Tryptophan Quenching Binding Assay
To study the binding properties of BcTSPO for PpIX, the tryptophan quenching binding assay was used as previously described with minor modifications.18,39 Detergent-solubilized BcTSPO, which exhibited the same catalytic ability as nanodisc-reconstituted BcTSPO (Figures S2 and S4C), was prepared and titrated with increasing amounts of PpIX. Briefly, purified protein in buffer D at a final concentration of 3.16 μM was titrated with increasing amounts of PpIX in the desired concentration range. Each titration point was followed by a spectrum measurement and detection of the intrinsic tryptophan fluorescence of BcTSPO. BcTSPO was excited at 280 nm, and emission was measured at 300–400 nm using the F-7000 FL spectrophotometer (Hitachi). The resulting data points were fitted with the following equation for dissociation constant Kd.
 |
where % quenching represents the calculated percentage quenching at each PpIX concentration, Fi is the maximal fluorescence intensity of BcTSPO tryptophan residues without PpIX, Ff is the maximal fluorescence intensity of BcTSPO tryptophan residues when mixed with 23.71 μM PpIX, and cTSPO and cPpIX are the concentrations of TSPO and PpIX ligands, respectively. The experiments were performed in triplicate and are shown as averages with standard error.
DEER Measurements and Analysis
DEER experiments were conducted at 80 K with a Bruker ELEXSYS E580-400 CW/pulsed spectrometer with a split-ring resonator (EN4118X-MS3) and a helium gas flow system (4118CF and 411A). The DEER experiments were performed using the typical four-pulse constant-time DEER sequence as previously described.24,40 Data analysis for DEER was first performed using Tikhonov regularization with the L-curve method.41,42 Briefly, background signals in the time-domain DEER traces were removed utilizing the DeerAnalysis program41 followed by the Tikhonov regularization analysis to obtain model-free distance distributions. The model-free results gave useful information about the number of major populations in the conformational ensemble that could possibly coexist with the experimental DEER data. Interspin distance distributions can be modeled as a sum of Gaussians, as previously demonstrated.43−46 The model-based analysis (i.e., Gaussian-based) and error estimations for DEER were then performed utilizing the program DD (version 7).43 The optimal number of Gaussians determined in the model-based analysis varied from 2 to 3. In DD, it quantifies the errors in the distance distribution in terms of a confidence band about the best-fit solution. This study presents the result in a confidence band of 2σ (95.4%), which reflects the influence of both the noise in the measured DEER data and the uncertainty in the estimate of the background correction. The confidence band was calculated from the full covariance matrix using the delta method.
Spin-Counting Measurements by CW-ESR
To determine the R1 labeling efficiency, the continuous wave (CW)-ESR technique was used to calculate the spin concentration of spin-labeled BcTSPO mutants. Several single- and double-cysteine variants were prepared for spin-counting study. Spin labeling was performed by the addition of a 25-fold molar excess of MTSSL (Enzo Life Sciences) for each cysteine from a 0.1 M stock solution in acetonitrile. The labeling reaction was allowed to proceed in the dark at 4 °C overnight to yield the side chain R1. Spin-labeled variants of BcTSPO were buffer-exchanged into buffer D to remove unreacted MTSSL and concentrated by centrifugation followed by the addition of glycerol (containing 10% (v/v) glycerol as a cryoprotectant). The sample was loaded into a capillary tube, and each spectrum was recorded with a magnetic width of 200 G, 1024 points, and 5 scans at 200 K. The ESR absorption-mode spectrum was obtained by integration and compared with tempol (1–4 mM) to calculate the labeling efficiency of individual BcTSPO variants, and the protein concentration was determined using absorption spectroscopy at 280 nm.
Kinetics of TSPO-Mediated PpIX Degradation
To study the kinetics of BcTSPO for PpIX degradation, the fluorescence-based assay was used as previously described.18 Purified BcTSPO (4.73 μM) in buffer D was mixed with PpIX in the desired concentration range (1.5–23.7 μM) and loaded into a 1 mm square quartz cuvette with a total volume of 300 μL. We used an F-7000 FL spectrophotometer (Hitachi) to monitor the time-dependent PpIX fluorescence intensity at 633 nm when excited at 405 nm. The first 120 s of the acquired data was fitted to a straight line, and the absolute value of the slope was proportional to the initial reaction rate. The initial reaction rate V0 was calculated with the following equation.
where S represents the slope of the linear fit and cPpIX is the concentration of the PpIX ligand. Data were then fitted using the Michaelis–Menten equation to determine KM and Vmax values of BcTSPO for PpIX degradation, and shown are averages with standard error (SE).
TSPO PEG-maleimide Labeling
Following purification of TSPO, PEG conjugation was performed by the addition of a 5-fold molar excess of 5 kDa polyethylene glycol (PEG) (Sigma-Aldrich) methyl ether maleimide (mPEG-mal) for each TSPO cysteine variant from a 0.01 M stock solution in buffer D. The conjugating reaction was allowed to proceed in the dark at 4 °C overnight to yield the PEGylated TSPO. After reaction, TSPO-PEG was buffer-exchanged into buffer D to remove unreacted mPEG-mal. The PEG-labeling efficiency of TSPO mutants was >90%, confirmed by SDS-PAGE with Coomassie blue staining (Figure S7C), and the protein concentration was determined using absorption spectroscopy at 280 nm.
Acknowledgments
This work was supported by grants (108-2113-M-007-029, 111-2731-M-007-001, 111-2113-M-007-015, and 112-2923-M-007-003) from the National Science and Technology Council of Taiwan and the Frontier Research Center on Fundamental and Applied Sciences of Matters at NTHU. All the ESR measurements were conducted in the Research Instrument Center of Taiwan located at NTHU.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacsau.3c00514.
(Figures S1 and S2) Experimental fluorescence spectra and the results of spectral analysis, (Figure S3) Arpeggio analysis outcome, (Figures S4 and S5) DEER experimental and analysis results, (Figure S6) CW-ESR spin-counting data and a comparison of TSPO structures, (Figure S7) fluorescence spectra related to the investigation of the opening between TM1 and TM2, (Figure S8) DEER data verifying the monomer/dimer state of TSPO in nanodiscs and the results of purified TSPO nanodiscs, and (Figure S9) data for estimating the TSPO incorporation efficiency into nanodiscs (PDF)
Author Contributions
P.-S.Y., C.-C.L., and Y.-W.C. conceived the experiments and analyzed the data. P.-S.Y. and Y.-S.L. conducted experiments. Y.-W.C. supervised the work. All authors wrote and edited the manuscript.
The authors declare that they have no competing interests. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
The authors declare no competing financial interest.
Notes
No unexpected or unusually high safety hazards were encountered.
Supplementary Material
References
- Li F.; Liu J.; Zheng Y.; Garavito R. M.; Ferguson-Miller S. Crystal Structures of Translocator Protein (TSPO) and Mutant Mimic of a Human Polymorphism. Science 2015, 347, 555–558. 10.1126/science.1260590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.; Kalathur R. C.; Liu Q.; Kloss B.; Bruni R.; Ginter C.; Kloppmann E.; Rost B.; Hendrickson W. A. Structure and Activity of Tryptophan-Rich TSPO Proteins. Science 2015, 347, 551–555. 10.1126/science.aaa1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaremko Ł.; Jaremko M.; Giller K.; Becker S.; Zweckstetter M. Structure of the Mitochondrial Translocator Protein in Complex with a Diagnostic Ligand. Science 2014, 343, 1363–1366. 10.1126/science.1248725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.; Liu J.; Liu N.; Kuhn L. A.; Garavito R. M.; Ferguson-Miller S. Translocator Protein 18 KDa (TSPO): An Old Protein with New Functions?. Biochemistry 2016, 55, 2821–2831. 10.1021/acs.biochem.6b00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiser C.; Montgomery B. L.; Ferguson-Miller S. TSPO Protein Binding Partners in Bacteria, Animals, and Plants. J. Bioenerg. Biomembr. 2021, 53, 463–487. 10.1007/s10863-021-09905-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A. W. U.; Montgomery B. L. Interdependence of Tetrapyrrole Metabolism, the Generation of Oxidative Stress and the Mitigative Oxidative Stress Response. Redox Biol. 2015, 4, 260–271. 10.1016/j.redox.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batoko H.; Veljanovski V.; Jurkiewicz P. Enigmatic Translocator Protein (TSPO) and Cellular Stress Regulation. Trends Biochem. Sci. 2015, 40, 497–503. 10.1016/j.tibs.2015.07.001. [DOI] [PubMed] [Google Scholar]
- Ammer L.-M.; Vollmann-Zwerenz A.; Ruf V.; Wetzel C. H.; Riemenschneider M. J.; Albert N. L.; Beckhove P.; Hau P. The Role of Translocator Protein TSPO in Hallmarks of Glioblastoma. Cancers 2020, 12, 2973. 10.3390/cancers12102973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert N. L.; Unterrainer M.; Fleischmann D. F.; Lindner S.; Vettermann F.; Brunegraf A.; Vomacka L.; Brendel M.; Wenter V.; Wetzel C.; Rupprecht R.; Tonn J.-C.; Belka C.; Bartenstein P.; Niyazi M. TSPO PET for Glioma Imaging Using the Novel Ligand 18F-GE-180: First Results in Patients with Glioblastoma. Eur. J. Nucl. Med. Mol. Imaging 2017, 44, 2230–2238. 10.1007/s00259-017-3799-9. [DOI] [PubMed] [Google Scholar]
- Tournier B. B.; Tsartsalis S.; Rigaud D.; Fossey C.; Cailly T.; Fabis F.; Pham T.; Grégoire M.-C.; Kövari E.; Moulin-Sallanon M.; Savioz A.; Millet P. TSPO and Amyloid Deposits in Sub-Regions of the Hippocampus in the 3xTgAD Mouse Model of Alzheimer’s Disease. Neurobiol. Dis. 2019, 121, 95–105. 10.1016/j.nbd.2018.09.022. [DOI] [PubMed] [Google Scholar]
- Gatliff J.; Campanella M. TSPO: Kaleidoscopic 18-KDa amid Biochemical Pharmacology, Control and Targeting of Mitochondria. Biochem. J. 2016, 473, 107–121. 10.1042/BJ20150899. [DOI] [PubMed] [Google Scholar]
- El Chemali L.; Akwa Y.; Massaad-Massade L. The Mitochondrial Translocator Protein (TSPO): A Key Multifunctional Molecule in the Nervous System. Biochem. J. 2022, 479, 1455–1466. 10.1042/BCJ20220050. [DOI] [PubMed] [Google Scholar]
- Lee Y.; Park Y.; Nam H.; Lee J.-W.; Yu S.-W. Translocator Protein (TSPO): The New Story of the Old Protein in Neuroinflammation. BMB Rep. 2020, 53, 20–27. 10.5483/BMBRep.2020.53.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y.; Cui M.; Ochs K.; Brendel M.; Strübing F. L.; Briel N.; Eckenweber F.; Zou C.; Banati R. B.; Liu G.-J.; Middleton R. J.; Rupprecht R.; Rudolph U.; Zeilhofer H. U.; Rammes G.; Herms J.; Dorostkar M. M. Long-Term Diazepam Treatment Enhances Microglial Spine Engulfment and Impairs Cognitive Performance via the Mitochondrial 18 KDa Translocator Protein (TSPO). Nat. Neurosci. 2022, 25, 317–329. 10.1038/s41593-022-01013-9. [DOI] [PubMed] [Google Scholar]
- Jurkiewicz P.; Senicourt L.; Ayeb H.; Lequin O.; Lacapere J.-J.; Batoko H. A Plant-Specific N-Terminal Extension Reveals Evolutionary Functional Divergence within Translocator Proteins. iScience 2020, 23, 100889 10.1016/j.isci.2020.100889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeliseev A. A.; Krueger K. E.; Kaplan S. A Mammalian Mitochondrial Drug Receptor Functions as a Bacterial “Oxygen” Sensor. Proc. Natl. Acad. Sci. U.S.A. 1997, 94, 5101–5106. 10.1073/pnas.94.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti A.; Owen D. R.; Grozeva D.; Rabiner E. A.; Matthews P. M.; Craddock N.; Young A. H. Bipolar Disorder Is Associated with the Rs6971 Polymorphism in the Gene Encoding 18 kDa Translocator Protein (TSPO). Psychoneuroendocrinology 2013, 38, 2826–2829. 10.1016/j.psyneuen.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginter C.; Kiburu I.; Boudker O. Chemical Catalysis by the Translocator Protein (18 KDa). Biochemistry 2013, 52, 3609–3611. 10.1021/bi400364z. [DOI] [PubMed] [Google Scholar]
- Busch A. W. U.; WareJoncas Z.; Montgomery B. L. Tryptophan-Rich Sensory Protein/Translocator Protein (TSPO) from Cyanobacterium Fremyella Diplosiphon Binds a Broad Range of Functionally Relevant Tetrapyrroles. Biochemistry 2017, 56, 73–84. 10.1021/acs.biochem.6b01019. [DOI] [PubMed] [Google Scholar]
- Vanhee C.; Zapotoczny G.; Masquelier D.; Ghislain M.; Batoko H. The Arabidopsis Multistress Regulator TSPO Is a Heme Binding Membrane Protein and a Potential Scavenger of Porphyrins via an Autophagy-Dependent Degradation Mechanism. Plant Cell 2011, 23, 785–805. 10.1105/tpc.110.081570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma A.; Nye J. S.; Snyder S. H. Porphyrins Are Endogenous Ligands for the Mitochondrial (Peripheral-Type) Benzodiazepine Receptor. Proc. Natl. Acad. Sci. U.S.A. 1987, 84, 2256–2260. 10.1073/pnas.84.8.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachar M.; Anderson K. E.; Ma X. Protoporphyrin IX: The Good, the Bad, and the Ugly. J. Pharmacol. Exp. Ther. 2016, 356, 267–275. 10.1124/jpet.115.228130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.-C.; Hung C.-L.; Yeh P.-S.; Li C.-E.; Chiang Y.-W. Doubly Spin-Labeled Nanodiscs to Improve Structural Determination of Membrane Proteins by ESR. RSC Adv. 2019, 9, 9014–9021. 10.1039/C9RA00896A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.-C.; Kao T.-Y.; Cheng C.-C.; Chiang Y.-W. Structure and Regulation of the BsYetJ Calcium Channel in Lipid Nanodiscs. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 30126–30134. 10.1073/pnas.2014094117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiemann O.; Heubach C. A.; Abdullin D.; Ackermann K.; Azarkh M.; Bagryanskaya E. G.; Drescher M.; Endeward B.; Freed J. H.; Galazzo L.; Goldfarb D.; Hett T.; Esteban Hofer L.; Fábregas Ibáñez L.; Hustedt E. J.; Kucher S.; Kuprov I.; Lovett J. E.; Meyer A.; Ruthstein S.; Saxena S.; Stoll S.; Timmel C. R.; Di Valentin M.; Mchaourab H. S.; Prisner T. F.; Bode B. E.; Bordignon E.; Bennati M.; Jeschke G. Benchmark Test and Guidelines for DEER/PELDOR Experiments on Nitroxide-Labeled Biomolecules. J. Am. Chem. Soc. 2021, 143, 17875–17890. 10.1021/jacs.1c07371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debruycker V.; Hutchin A.; Masureel M.; Ficici E.; Martens C.; Legrand P.; Stein R. A.; Mchaourab H. S.; Faraldo-Gómez J. D.; Remaut H.; Govaerts C. An Embedded Lipid in the Multidrug Transporter LmrP Suggests a Mechanism for Polyspecificity. Nat. Struct. Mol. Biol. 2020, 27, 829–835. 10.1038/s41594-020-0464-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T.; Wang D.; Jeon J.; Schwieters C. D.; Clore G. M. Quantitative Agreement between Conformational Substates of Holo Calcium-Loaded Calmodulin Detected by Double Electron–Electron Resonance EPR and Predicted by Molecular Dynamics Simulations. J. Am. Chem. Soc. 2022, 144, 12043–12051. 10.1021/jacs.2c02201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastvan R.; Mishra S.; Peskova Y. B.; Nakamoto R. K.; Mchaourab H. S. Mechanism of Allosteric Modulation of P-Glycoprotein by Transport Substrates and Inhibitors. Science 2019, 364, 689–692. 10.1126/science.aav9406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubb H. C.; Higueruelo A. P.; Ochoa-Montaño B.; Pitt W. R.; Ascher D. B.; Blundell T. L. Arpeggio: A Web Server for Calculating and Visualising Interatomic Interactions in Protein Structures. J. Mol. Biol. 2017, 429, 365–371. 10.1016/j.jmb.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagelueken G.; Abdullin D.; Schiemann O. MtsslSuite: Probing Biomolecular Conformation by Spin-Labeling Studies. Methods Enzymol. 2015, 563, 595–622. 10.1016/bs.mie.2015.06.006. [DOI] [PubMed] [Google Scholar]
- Xu Y.; Wang S.; Hu Q.; Gao S.; Ma X.; Zhang W.; Shen Y.; Chen F.; Lai L.; Pei J. CavityPlus: A Web Server for Protein Cavity Detection with Pharmacophore Modelling, Allosteric Site Identification and Covalent Ligand Binding Ability Prediction. Nucleic Acids Res. 2018, 46, W374–W379. 10.1093/nar/gky380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Hiser C.; Li F.; Hall R.; Garavito R. M.; Ferguson-Miller S. New TSPO Crystal Structures of Mutant and Heme-Bound Forms with Altered Flexibility, Ligand Binding, and Porphyrin Degradation Activity. Biochemistry 2023, 62, 1262–1273. 10.1021/acs.biochem.2c00612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurrus E.; Engel D.; Star K.; Monson K.; Brandi J.; Felberg L. E.; Brookes D. H.; Wilson L.; Chen J.; Liles K.; Chun M.; Li P.; Gohara D. W.; Dolinsky T.; Konecny R.; Koes D. R.; Nielsen J. E.; Head-Gordon T.; Geng W.; Krasny R.; Wei G.; Holst M. J.; McCammon J. A.; Baker N. A. Improvements to the APBS Biomolecular Solvation Software Suite. Protein Sci. 2018, 27, 112–128. 10.1002/pro.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens C.; Stein R. A.; Masureel M.; Roth A.; Mishra S.; Dawaliby R.; Konijnenberg A.; Sobott F.; Govaerts C.; Mchaourab H. S. Lipids Modulate the Conformational Dynamics of a Secondary Multidrug Transporter. Nat. Struct. Mol. Biol. 2016, 23, 744–751. 10.1038/nsmb.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho P.-S.; Kao T.-Y.; Li C.-C.; Lan Y.-J.; Lai Y.-C.; Chiang Y.-W. Nanodisc Lipids Exhibit Singular Behaviors Implying Critical Phenomena. Langmuir 2022, 38, 15372–15383. 10.1021/acs.langmuir.2c02596. [DOI] [PubMed] [Google Scholar]
- Denisov I. G.; Grinkova Y. V.; Lazarides A. A.; Sligar S. G. Directed Self-Assembly of Monodisperse Phospholipid Bilayer Nanodiscs with Controlled Size. J. Am. Chem. Soc. 2004, 126, 3477–3487. 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- Bayburt T. H.; Grinkova Y. V.; Sligar S. G. Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins. Nano Lett. 2002, 2, 853–856. 10.1021/nl025623k. [DOI] [Google Scholar]
- Marsh D.Handbook of Lipid Bilayers, 2nd ed.; CRC Press, Taylor & Francis Group: Boca Raton, FL, 2013 10.1201/b11712. [DOI] [Google Scholar]
- Li F.; Xia Y.; Meiler J.; Ferguson-Miller S. Characterization and Modeling of the Oligomeric State and Ligand Binding Behavior of Purified Translocator Protein 18 KDa from Rhodobacter Sphaeroides. Biochemistry 2013, 52, 5884–5899. 10.1021/bi400431t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeschke G. DEER Distance Measurements on Proteins. Annu. Rev. Phys. Chem. 2012, 63, 419–446. 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- Jeschke G.; Chechik V.; Ionita P.; Godt A.; Zimmermann H.; Banham J.; Timmel C. R.; Hilger D.; Jung H. DeerAnalysis2006—a Comprehensive Software Package for Analyzing Pulsed ELDOR Data. Appl. Magn. Reson. 2006, 30, 473–498. 10.1007/BF03166213. [DOI] [Google Scholar]
- Chiang Y.-W.; Borbat P. P.; Freed J. H. The Determination of Pair Distance Distributions by Pulsed ESR Using Tikhonov Regularization. J. Magn. Reson. 2005, 172, 279–295. 10.1016/j.jmr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Stein R. A.; Beth A. H.; Hustedt E. J. A Straightforward Approach to the Analysis of Double Electron–Electron Resonance Data. Methods Enzymol. 2015, 563, 531–567. 10.1016/bs.mie.2015.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hustedt E. J.; Marinelli F.; Stein R. A.; Faraldo-Gómez J. D.; Mchaourab H. S. Confidence Analysis of DEER Data and Its Structural Interpretation with Ensemble-Biased Metadynamics. Biophys. J. 2018, 115, 1200–1216. 10.1016/j.bpj.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung T.-C.; Li C.-Y.; Lai Y.-C.; Hung C.-L.; Shih O.; Yeh Y.-Q.; Jeng U.-S.; Chiang Y.-W. Solution Structure of Apoptotic BAX Oligomer: Oligomerization Likely Precedes Membrane Insertion. Structure 2015, 23, 1878–1888. 10.1016/j.str.2015.07.013. [DOI] [PubMed] [Google Scholar]
- Brandon S.; Beth A. H.; Hustedt E. J. The Global Analysis of DEER Data. J. Magn. Reson. 2012, 218, 93–104. 10.1016/j.jmr.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.