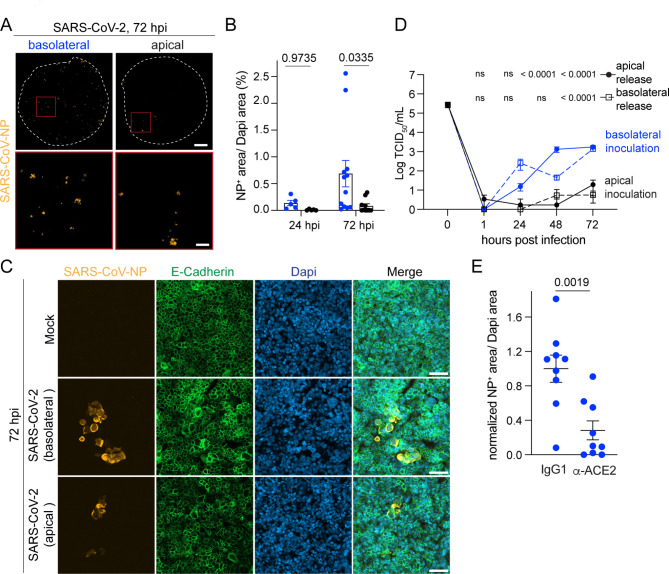
Fig. 4.
SARS-CoV-2 infects HIBCPP preferentially from the basolateral side. (A) Representative tile stack images from immunofluorescence staining for SARS-CoV-nucleocapsid protein (NP) (orange) of whole filter inserts with HIBCPP cells 72 hpi with SARS-CoV-2 at 100’000 TCID50/filter insert from basolateral and apical side are shown. White dashed circle indicates border of the filter inserts, red box delineates inset. Scale bar = 1 mm and for inset 200 μm. (B) Quantification of NP+ area normalized to DAPI+ area 24 and 72 hpi with SARS-CoV-2. Each dot represents a replicate from in total four experiments. P-values were determined by two-way Anova followed by Sidàk test to correct for multiple comparison. (C) Representative confocal images from immunofluorescence staining of SARS-CoV-NP (orange) and E-Cadherin (green) 72 hpi with SARS-CoV-2 from basolateral and apical side or Mock are shown. Nuclei were stained with DAPI (blue). Scale bar = 50 μm. (D) Quantification of released virions into the basolateral and apical supernatant 1–72 hpi with SARS-CoV-2 by TCID50 assay. Measurement was done in duplicates or triplicates from a total of 4 experiments. P-values were determined using 2-way Anova followed by Tukey’s test to correct for multiple comparison and are indicated for the comparison of the supernatant from the respective compartment from basolateral versus apical infection. (E) Quantification of NP+ area normalized to DAPI+ area 72 hpi with SARS-CoV-2 at 100’000 TCID50/filter insert from basolateral side in presence of anti-ACE2 blocking or IgG control antibody. Data is normalized to IgG control. Each dot represents a replicate from in total 3 experiments. P-value was calculated using unpaired T-test with Welch’s correction