Figure 1.
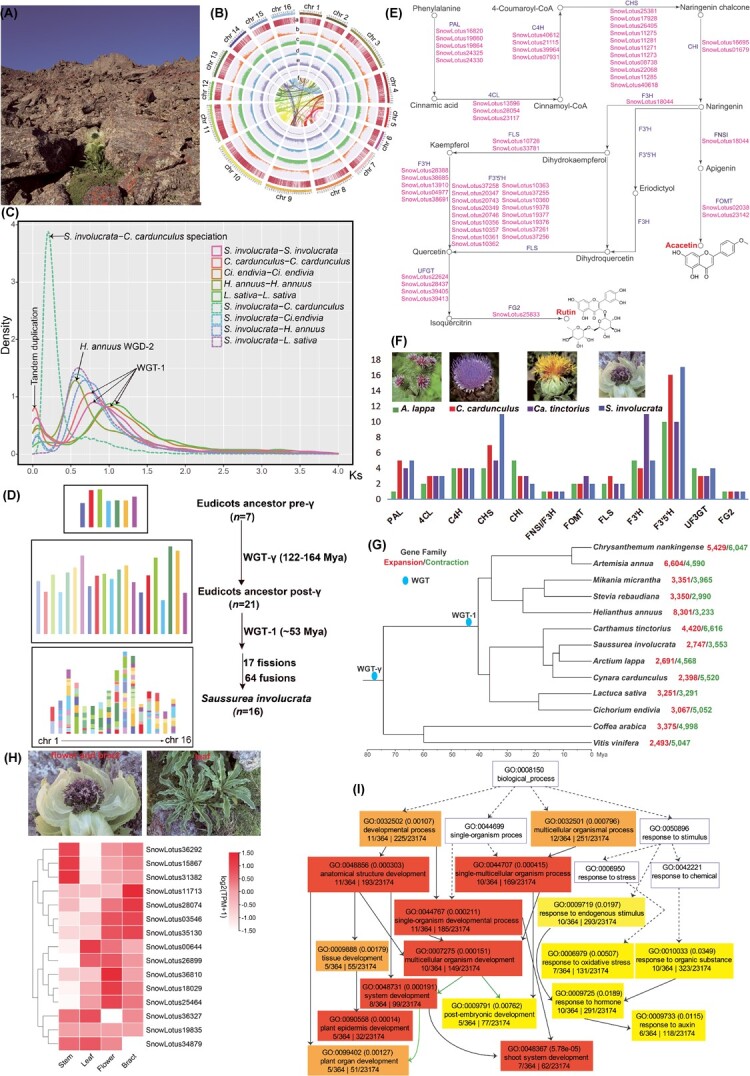
Assembly, annotation, and analyses of the Saussurea involucrata genome. All S. involucrata photographs are taken by Dr. Bing Liu from the Institute of Botany, Chinese Academy of Sciences. (A) Photographs showing habitat of S. involucrata. (B) Genomic landscape of S. involucrata. (f) GC content, (e) Gypsy LTR density, (d) Copia LTR density, (c) total LTR density, (b) gene density, and (a) gene expression in leaf. (C) Distribution of synonymous substitution (Ks) of S. involucrata paralogues, Cynara cardunculus paralogues, Cichorium endivia paralogues, Helianthus annuus paralogues, Lactuca sativa paralogues, S. involucrata-C. cardunculus orthologues, S. involucrata-Ci. endivia orthologues, S. involucrata-H. annuus orthologues, and S. involucrata-L. sativa orthologues. (D) Evolutionary scenario of S. involucrata from the ancestral eudicot karyotype. (E) Predicted candidate genes involved in acacetin and rutin biosynthesis in S. involucrata. (F) Comparison of gene numbers in gene families involved in acacetin and rutin biosynthesis between S. involucrata and three other Asteraceae species. The images of C. cardunculus, Arctium lappa, and Carthamus tinctorius are from Plant Photo Bank of China (PPBC). (G) Phylogenetic analysis among 13 eudicot species, including S. involucrata and 10 other Asteraceae species, with information of expansion/contraction of gene families. (H) Expression profiles of identified genes related to cold resistance in S. involucrata across four tissues: stem, leaf, flower, and bract. (I) Enriched GO terms (highlighted with colors) of genes specifically expressed in S. involucrata bracts.
