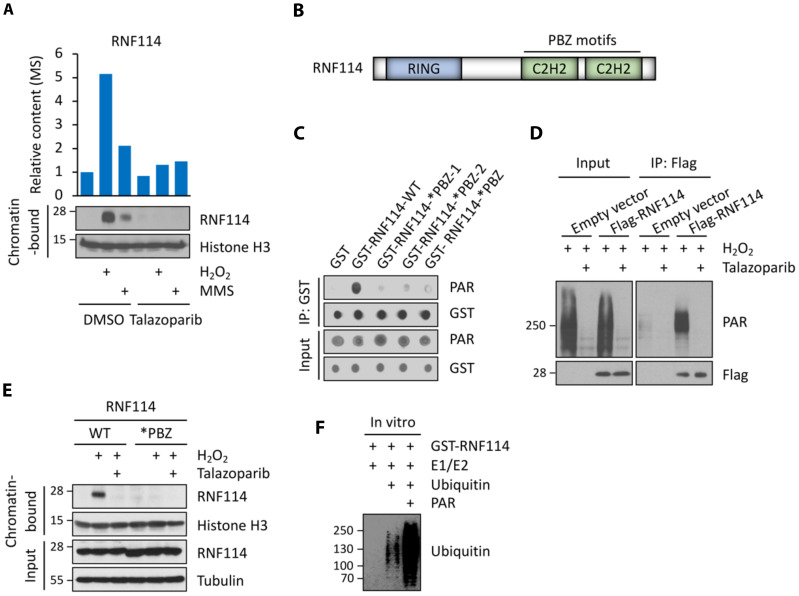
Fig. 2. Identification of RNF114 as a PAR-binding, E3 ubiquitin ligase.
(A) Abundances of RNF114 in the chromatin fraction as determined from the chromatin relocalization screen (top) and biochemical assays (bottom). (B) Domain structure of RNF114. (C and D) RNF114 binds to PAR polymers in vitro (C) and in cells (D). (C) Both RNF114-WT and the various RNF114 mutants were analyzed using the PAR dot blot assay (i.e., RNF114-WT; RNF114-*PBZ-1, C143A/C146A; RNF114-*PBZ-2, C173A/C176A; RNF114-*PBZ, C143A/C146A/C173A/C176A). (D) HCT116 cells expressing the empty vector or Flag-RNF114 were pretreated with talazoparib (1 μM for 1 hour) and then were treated with H2O2 (2 mM for 5 min). Cell lysates were subject to immunoprecipitation (IP) using an anti-FLAG antibody, and the immunoprecipitants were analyzed using the indicated antibodies. (E) PAR binding mediates the translocation of RNF114 to chromatin in response to genotoxic stress. RNF114-KO HCT116 cells were reconstituted with either RNF114-WT or RNF114-*PBZ mutant (the PAR-binding mutant). These cells were pretreated with talazoparib (1 μM for 1 hour) and then were treated with H2O2 (2 mM for 5 min). The chromatin-bound fraction was isolated from these cells and was subjected to immunoblotting experiments using the indicated antibodies. (F) PAR polymers stimulate the E3 activity of RNF114. Recombinant RNF114 was subject to in vitro ubiquitination experiments in the presence (0.4 μM) or absence of added PAR polymers. Immunoblot experiments were used to analyze RNF114 auto-ubiquitination.