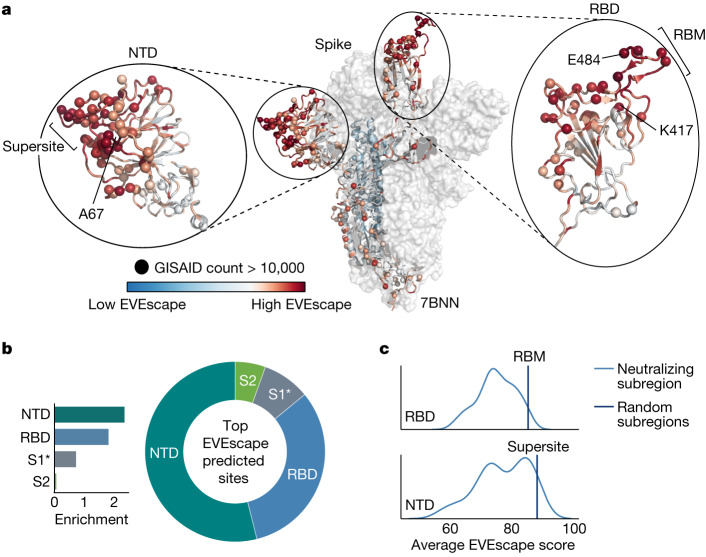
Fig. 2. EVEscape identifies antigenic regions without antibody information.
a, EVEscape scores (site-level maximum) mapped onto a representative Spike three-dimensional structure (Protein Data Bank (PDB) identifier: 7BNN) highlight high-scoring regions with many observed pandemic variants, both in the RBD and in the NTD. Spheres indicate sites with a total number of mutations observed more than 10,000 times in the GISAID sequence database. b, The top decile of EVEscape predictions span diverse epitope regions across Spike, but most of the predictions are in the NTD and RBD, which have a disproportionately high number of predicted EVEscape sites relative to their sequence length (enrichment). The regions considered are NTD (sequence positions 14–306), RBD (319–542), S1* (543–685) and S2 (686–1273), where S1* refers to the region in S1 between RBD and the S2. c, Neutralizing subregions—RBM (receptor-binding motif, 438–506) and NTD supersite36 (14–20,140–158, 245–263)—have significantly higher than average EVEscape scores, relative to a distribution of 150 random contiguous regions of the same length within the RBD and NTD, respectively.