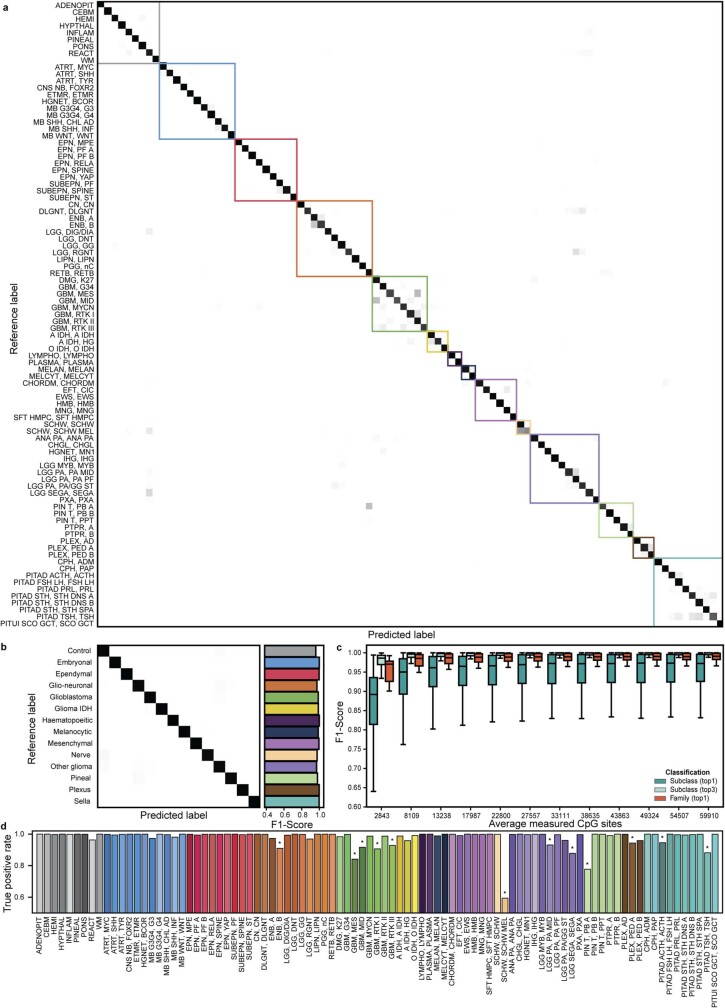
Extended Data Fig. 2. Sturgeon performance at 40 min of simulated sequencing.
a, Confusion matrix showing the highest scoring class for each reference label at 40 min of simulated sequencing (∼97% missing values from microarray data) b, Confusion matrix and F1 scores at 40 min of simulated sequencing when scores are aggregated on the family level. c, F1 scores at different sequencing depths (represented by the average number of covered 450 K array methylation sites) when classifying by subclass, by the correct subclass being in the top 3 of highest scoring classes and at the family level. Box plot minimum and maximum bounds represent the 25th and 75th percentiles, respectively, and the center bound represents the median. Whiskers extend to 1.5 times the interquartile range d, True positive rate for each subclass at 40 min of sequencing at the 0.95 confidence threshold. Asterisks indicate subclasses that do not reach the 0.95 true positive rate.