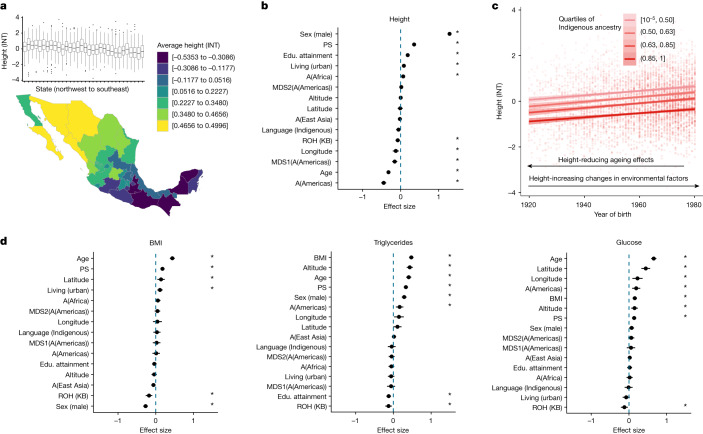
Fig. 5. An analysis of the factors influencing height and other complex trait variation.
a, Bottom: map of average height in Mexico (n = 5,770). Height was normalized using an INT. Top: box plots of height (INT) variation in each state from northwest to southeast. The box plots show the median value and the quartiles. Whiskers extend to the minimum and the maximum values. The dots represent outliers. n = 5,846 biologically independent samples were used for the analysis. b, Explanatory model for height variation implicates the role of genetics and environment. The plot shows effect-size estimates and confidence intervals (1.96 × s.e.m.) from a mixed-model analysis. All quantitative predictors are centred and scaled by 2 standard deviations. Asterisks show significance at false discovery rate < 0.05 across traits and predictors analysed50. n = 4,625 biologically independent samples were used for the analysis. c, Height as a function of birth year in quartiles of ancestries from the Americas (n = 5,598). Error bands represent 95% confidence intervals. d, Trait profiles for BMI (left), triglycerides (middle) and glucose (right). Results of mixed-model analysis, as in b. The plot shows effect-size estimates and confidence intervals (1.96 × s.e.m.) from a mixed-model analysis. n = 4,607, 3,664 and 3,613 biologically independent samples were used for the analysis for BMI, triglycerides and glucose, respectively. For b and d, PS are polygenic scores computed using UKB summary statistics (SNPs significant at P < 10–8), A(Africa/East Asia/Americas) refers to ancestry proportions from that region as inferred from ADMIXTURE, and MDS1(A(Americas)) and MDS2(A(Americas)) refers to multidimensional scaling (MDS) axes within ancestries from the Americas as inferred using a MAAS-MDS analysis (Supplementary Fig. 24). Educational (Edu.) attainment is on a scale from 0 to 8 (low to high educational attainment), and altitude is measured in metres (low to high).