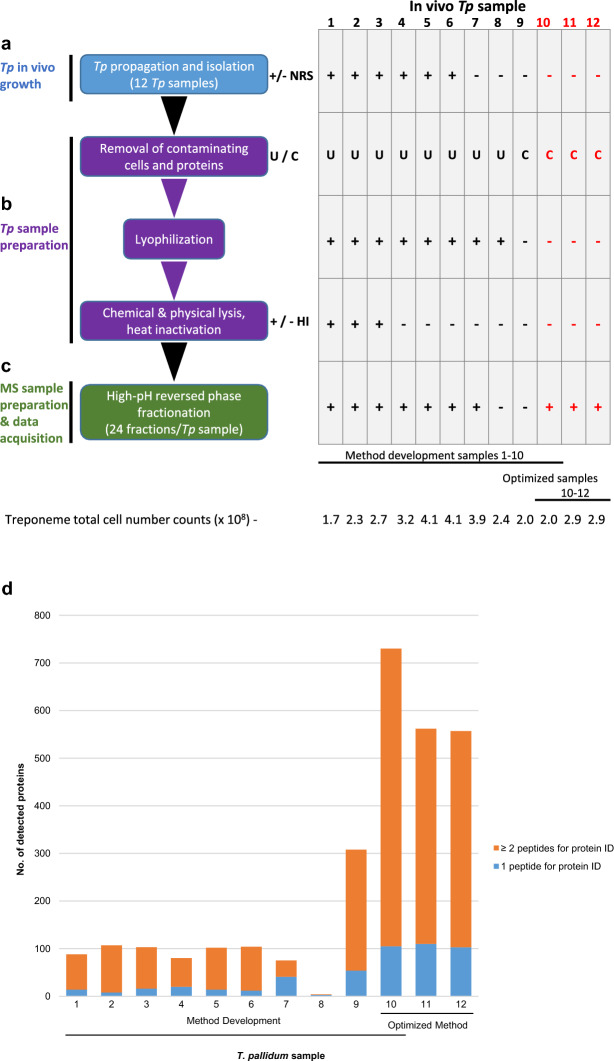
Figure 1.
Optimization of a workflow for deep proteome coverage of in vivo-grown T. pallidum. Schematics showing (a) isolation of T. pallidum from rabbits, (b) the major steps comprising T. pallidum sample preparation, and (c) the main optimization step in mass spectrometry sample preparation, processing, and data acquisition. The workflow indicates each of the individual steps that were performed in the optimization of the protocol for global profiling of T. pallidum protein expression (left). The corresponding table shows the variable parameter conditions used in each of the biological replicate samples at each of the individual optimization steps (right). Samples 1–10 were used for optimizing the protocol during the method development stages; sample 10 conditions were found to be optimal (red text). Samples 11 and 12 (red text) correspond to two biological replicate samples that were processed using the optimized protocol used for sample 10 to obtain three biological replicate samples prepared using an identical protocol. The total number of treponemes used in the preparation of each of the 12 samples is indicated (bottom). Plus sign; a parameter condition has been included in a protocol step; minus sign, a parameter condition has been omitted in a protocol step. +/− NRS = the addition or omission of normal rabbit serum (NRS) during T. pallidum isolation. U/C = the use of either ultrafiltration (U) or high-speed centrifugation (C) during removal of contaminating rabbit components. +/− HI = the use or omission of heat inactivation during T. pallidum sample preparation. (d) The total number of T. pallidum proteins that were detected and identified in each of the 12 in-vivo grown T. pallidum biological replicate samples. For each bar, the number of T. pallidum proteins that were identified via the detection of one tryptic peptide (blue) or two tryptic peptides (orange) in each of the 12 individual samples is indicated.