Figure 1.
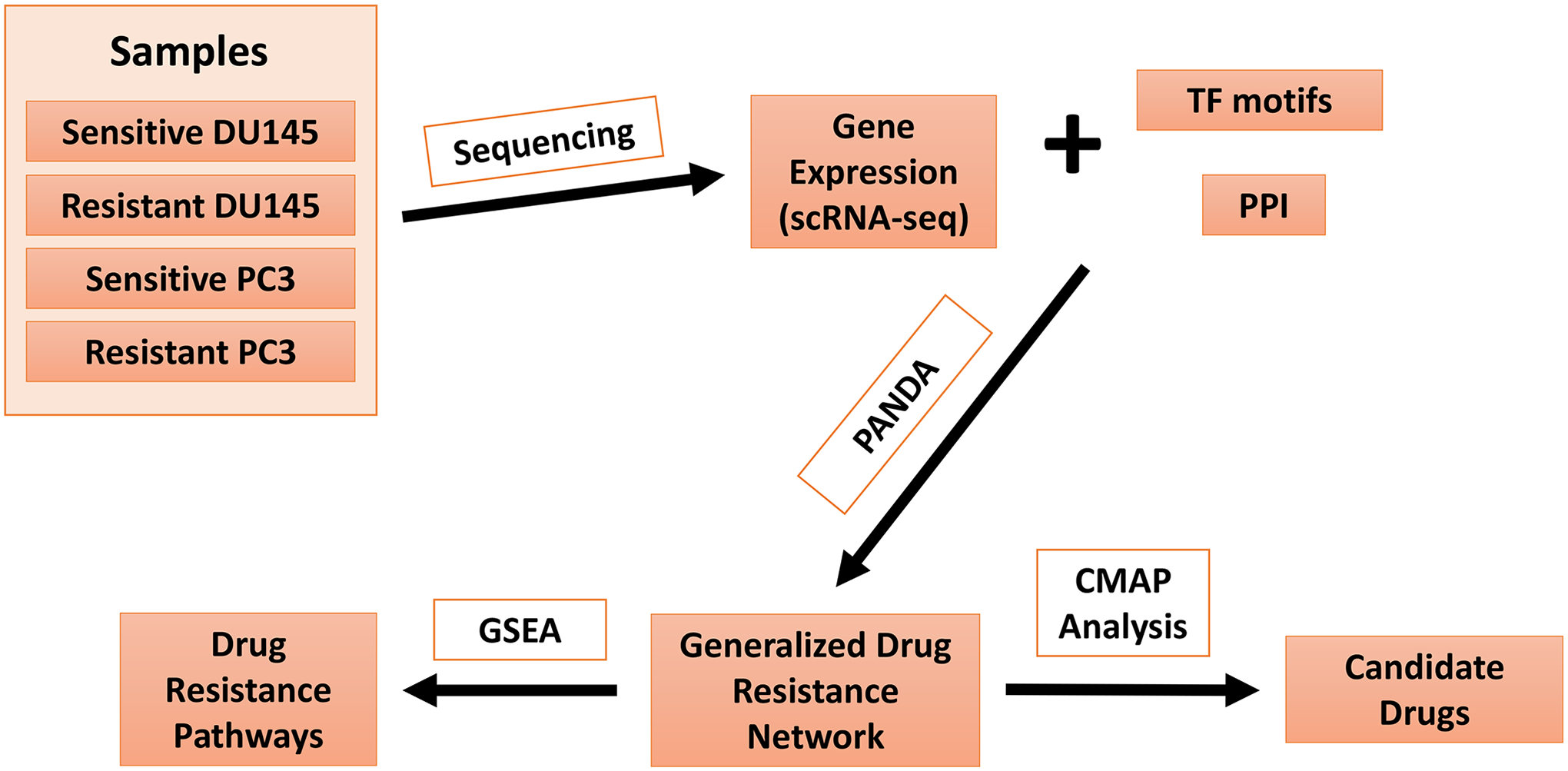
General Network Analysis Pipeline. Sensitive and resistant samples are subjected to scRNA-seq, followed by subjecting the scRNA-seq data in combination with transcription factor (TF) motifs and protein-protein interactions (PPI), derived from established databases, to PANDA analysis. This will lead to identifications of a generalized drug resistance network which can then be subjected to gene set enrichment analysis (GSEA) and connectivity map analysis (CMAP) to identify drug resistance pathways and candidate drugs, respectively.
