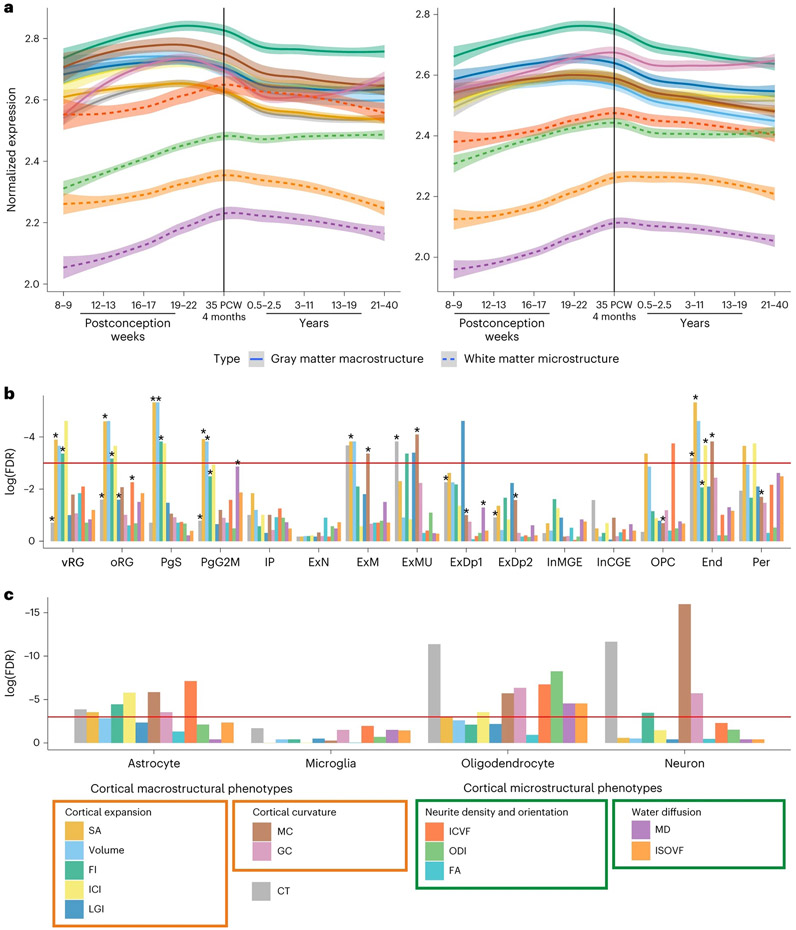
Fig. 4 ∣. Enrichment of GWAS signals in different cell types during development.
a, Developmental trajectories of average gene expression in cortical postmortem-bulk RNA data (PsychEncode) for all significant genes (FDR < 0.05, n = 34–1,113; Supplementary Table 12) identified using H-MAGMA (left) or MAGMA (right) for 12 of the 13 global phenotypes. Data for FA are not shown as too few genes were identified as significant. The shaded region indicates 95% confidence intervals. b, Results of enrichment analyses for cell-specific gene expression from midgestation. FDR-corrected log10(P values) for gene enrichment using genes identified from MAGMA (multiple regression) are plotted. The red line indicates the significance threshold at FDR-corrected P ≤ 0.05. Additionally, significant enrichments identified using H-MAGMA genes are indicated with an asterisk. c, Results of enrichment analyses from cell-specific epigenetic marks from postnatal cortex identified using LDSC-based enrichment. End, endothelial cells; ExDp1, excitatory deep layer neurons 1; ExDp2, excitatory deep layer neurons 2; ExM, maturing excitatory neurons; ExN, migrating excitatory neurons; ExMU, maturing excitatory neuron, upper enriched; InCGE, CGE interneuron; InMGE, MGE interneuron; IP, intermediate progenitors; OPC, oligodendrocyte precursor cells; oRG, outer radial glia; PgS and PgG2M, cycling progenitors, S phase and G2-M phase, respectively; Per, pericytes; vRG, ventral radial glia.