Figure 6. prdm16 and mecom are required for normal muscle innervation.
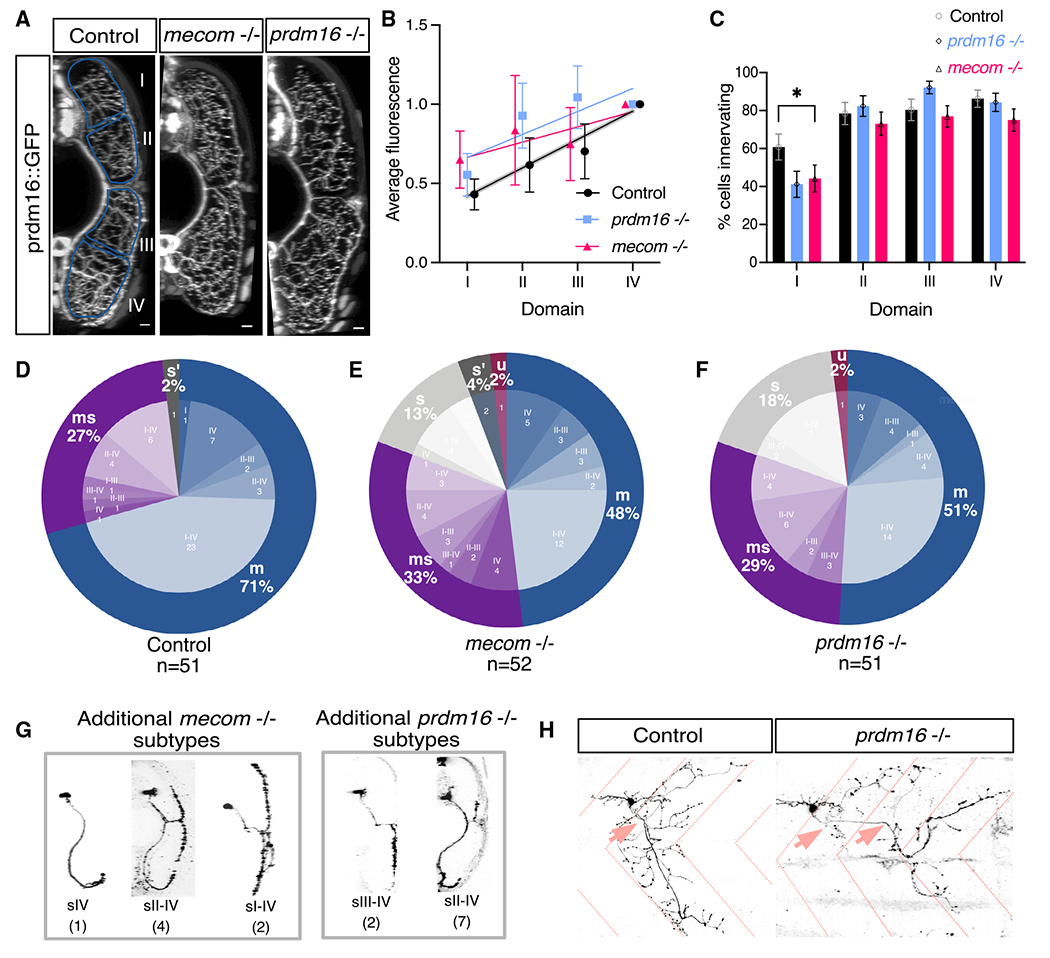
(A) Transverse view of the zebrafish tail showing Prdm16+ MN muscle innervation in Tg(prdm16::GFP) control, mecom mutant, and prdm16 mutant larvae at 5 dpf. Scale bars, 10 μm.
(B) Average GFP fluorescence of each muscle quadrant normalized to the fourth quadrant for control Tg(prdm16::GFP) fish (n = 6), mecom mutant (n = 9), and prdm16 mutant (n = 10) larvae at 5 dpf. Error bars indicate SEM. Lines illustrate linear regression for each condition and gray area surrounding control indicates SD of the slope, ±0.00555.
(C) Percentage of cells from control, mecom mutant, and prdm16 mutant Tg(prdm16:GAL4) sparse label experiments in (D)–(F) that innervated each dorsoventral muscle quadrant. Asterisk indicates significant difference (prdm16 −/− domain I, p = 0.000891; mecom −/− domain I, p = 0.004121), all other domains were not significantly different (p > 0.00625).
(D) Pie chart showing subtype proportions labeled in Tg(prdm16:GAL4) sparse label for control larvae. Replicated from Figure 5E.
(E) Pie chart showing subtype proportions labeled in Tg(prdm16:GAL4) sparse label for mecom mutant larvae.
(F) Pie chart showing subtype proportions labeled in Tg(prdm16:GAL4) sparse label for prdm16 mutant larvae.
(G) Example images of all sparsely labeled subtypes unique to mutants (E and F). Number in parentheses indicates number of times subtype was labeled.
(H) Example of an unclassifiable cell found in mutant sparse label experiment from the lateral view (right) compared with a control cell (left). Dashed coral lines indicate myotome boundaries. Coral arrows indicate motor exit points. Controls can indicate either WT or prdm16 +/− or mecom +/− animals, which are phenotypically indistinguishable.
