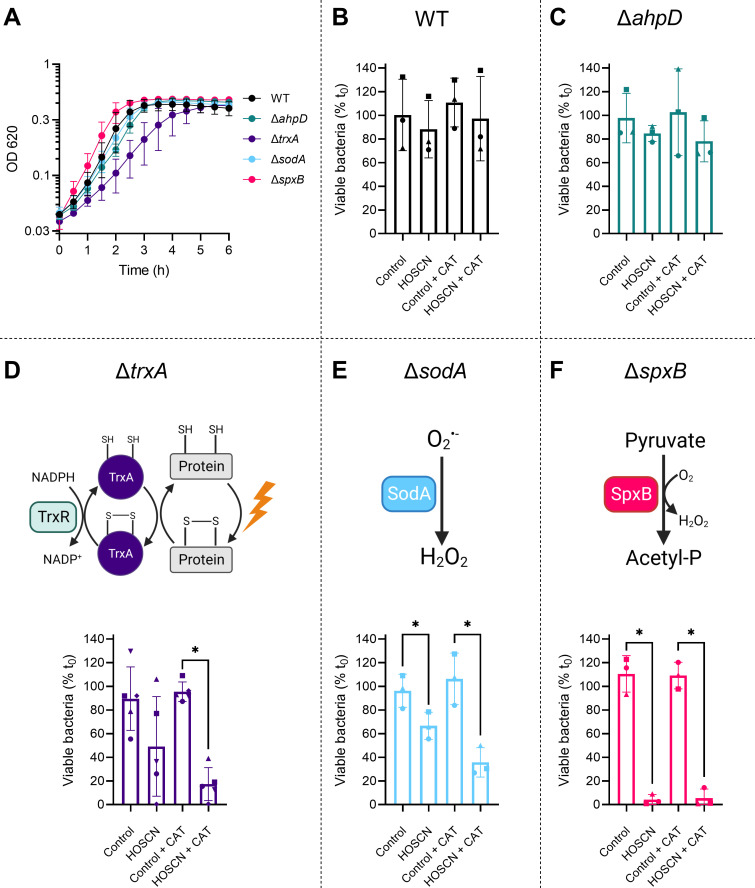
Fig 3.
Validation of Tn-seq using single-gene deletion mutants. (A) S. pneumoniae WT and mutant strains were grown in a 96-well plate in brain heart infusion media for 6 hours at 37°C in the presence of 5% CO2. Absorbance measurements (OD620) were taken every 30 min, following which the media-alone sample values were subtracted. Data are presented as the mean ± SD from at least three independent experiments, the y-axis is log10 scale. (B–F) S. pneumoniae strains (B) WT, (C) ΔahpD, (D) ΔtrxA, (E) ΔsodA, and (F) ΔspxB (OD620 0.02) were incubated in the presence or absence of 800 µM HOSCN ± catalase (CAT, 20 µg/mL) for 90 min in Hank’s balanced salt solution buffer, pH 6.8. Viable bacteria were determined by counting colony forming units and expressed relative to those before incubation (t 0). Data are presented as the mean ± SD from at least three independent experiments, with symbols designating matching replicate data. A significant difference to the respective control was determined by paired t-tests and is indicated by * for P < 0.05. (D–F) Schematic representation of the function of proteins TrxA, SodA, and SpxB, in reducing oxidized thiols in proteins, detoxifying superoxide, and metabolizing pyruvate, respectively. The schematic representations were created with Biorender.com.