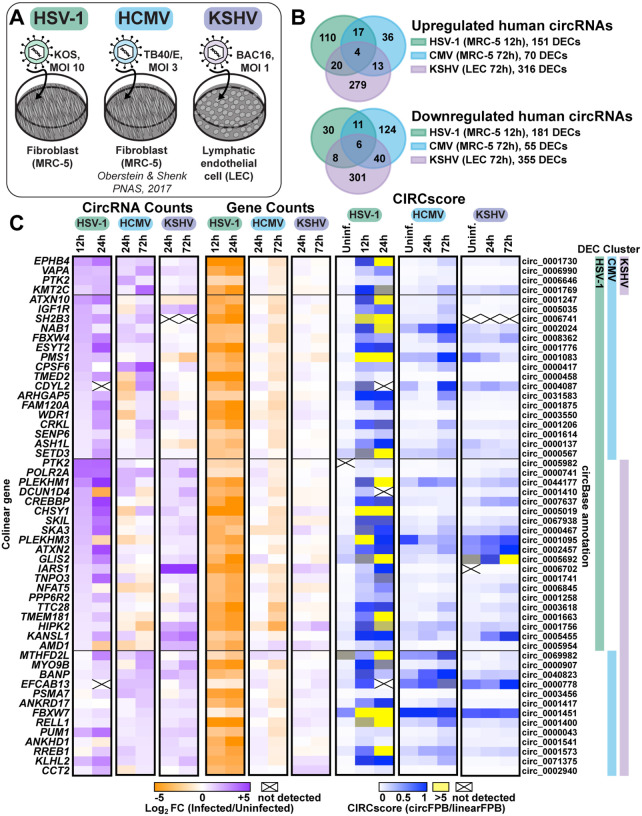
Figure 1. Human circRNAs upregulated in de novo lytic infection models.
A) Infographic for infection models used in this study. For HSV-1, fibroblasts (MRC-5) were infected with strain KOS at a multiplicity of infection (MOI) of 10 for 12 hours. For HCMV, fibroblasts (MRC-5) were infected with strain TB40/E at an MOI of 3 for 72 hours (Oberstein & Shenk 2017). For KSHV, human dermal lymphatic endothelial cells (LEC) were infected with strain BAC16 at an MOI of 1 for 72 hours. B) Overlap of differentially expressed circRNAs (DECs) detected by bulk RNA-Seq from HSV-1 (n=2–4), HCMV (n=2), and KSHV (n=2) infection. DECs had a raw back splice junction (BSJ) count across the sample set >10, Log2FC (fold change)>0.5 or <-0.5, and rank product p-value <0.05. C) Heatmaps for DECs which overlap between viruses, with DEC clusters indicating which virus the circRNA was found to be significantly upregulated within. Data is plotted as CircRNA counts (Log2FC Infected/Uninfected normalized BSJ counts), Gene counts (Log2FC Infected/Uninfected normalized gene counts), or CIRCscore (circFPB (fragments per billion mapped bases)/linearFPB). Heatmap values are the average of biological replicates. Log2FC is relative to a paired uninfected control.