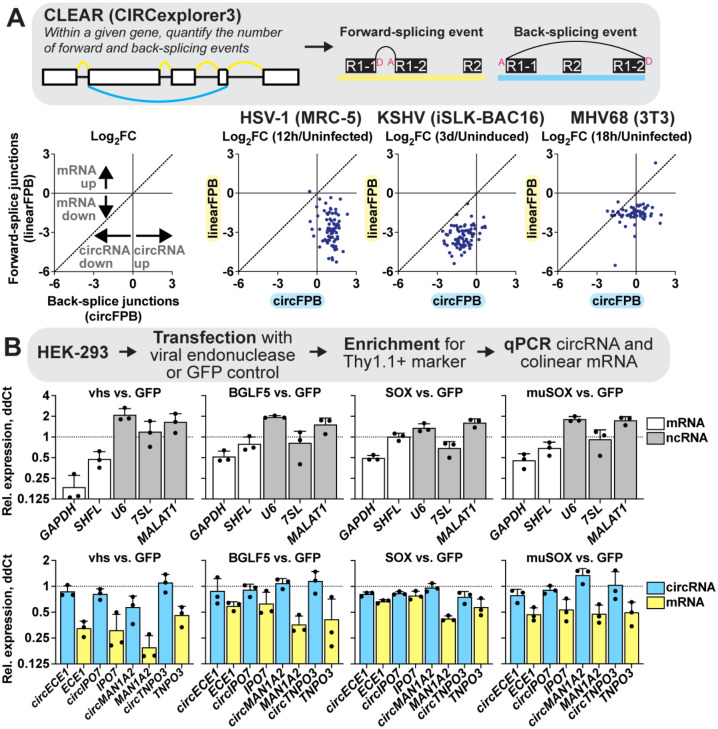
Figure 3. CircRNA are resistant to viral endonuclease mediated decay.
A) Bulk RNA-Seq analyzed using CLEAR (CircExplorer3) from HSV-1 (MRC-5 infected with KOS MOI 10 for 12 hours, n=4), KSHV (iSLK-BAC16 reactivated with Dox/NaB for 3 days, n=4), and MHV68 (3T3 infected with H2B-YFP MOI 5 for 18 hours, n=2) infection. Graphs are limited to genes where raw BSJ and forward splice junction (FSJ) counts were >1 across all biological replicates. The average Log2FC Infected/Uninfected (HSV-1, MHV68) or Induced/Uninduced (KSHV) was plotted for linearFPB and circFPB, with each dot being a distinct gene. B) HEK-293 cells were transfected for 24 hours with plasmid vectors expressing GFP or viral endonucleases (vhs, BGLF4, SOX, muSOX). RNA was collected and assessed after reverse transcription using qPCR to quantitate mRNAs and noncoding RNAs (ncRNAs). Data is plotted as relative expression (ddCt) using 18S rRNA as the reference gene, and relative to a paired GFP transfected sample. Data points are biological replicates, column bars are the average, and error bars are standard deviation.