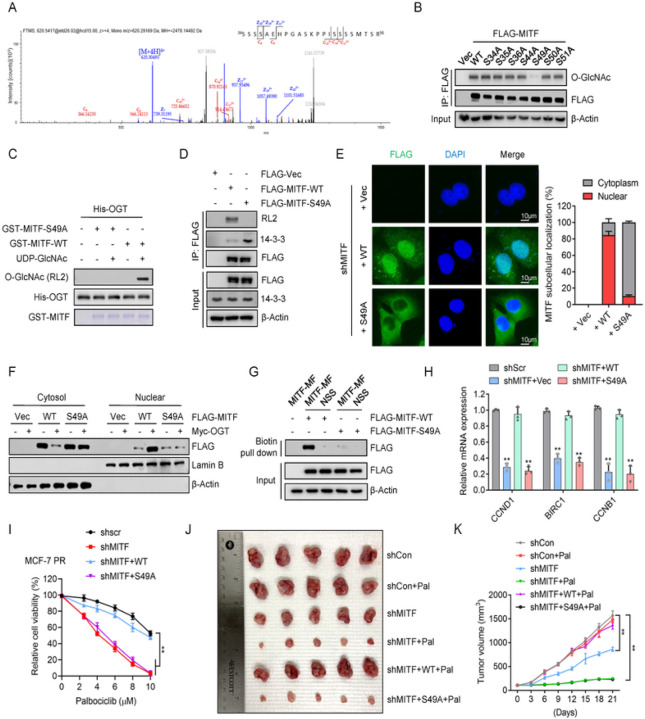
Figure 4. O-GlcNAcylation of MITF at S49 is required for its nuclear accumulation.
(A) MITF was purified from HEK293T cells and analyzed by LC-MS/MS analysis to identify the potential O-GlcNAcylation sites. The peptide of MITF from amino acids 34 to 56 with O-GlcNAcylation identified by LC-MS/MS was shown (note: S49 is a potential O-GlcNAc site). (B) HEK293T cells expressing indicated MITF or its O-GlcNAc mutants from plasmids were harvested48 hr after transfection, followed by co-IP and FLAG-IPs were then immunoblotted for indicated proteins. (C) Identification of the O-GlcNAcylation modification of MITF by in vitro O-GlcNAcylation assays. Recombinant GST-MITF, GST-MITF-S49A, and His-OGT were purified from E. coli. (D) MCF-7 PR cells were transfected with indicated plasmids for 48 hr before harvested for co-IP. FLAG-IPs were then immunoblotted for indicated proteins. (E) Immunofluorescence to examine the MITF localization with indicated treatment in MCF-7 PR cells. The scale bar represents 10 μm. Right, the quantification of results is shown on the left. (F) MCF-7 PR cells were transfected with indicated plasmids for 48 hr, then cytoplasmic and nuclear fractions were extracted and subjected to immunoblotting for indicated proteins. (G) MCF-7 PR cells were transfected with the indicated plasmids for 48 hr, and then the nuclear fraction of cell lysates was mixed with biotin-tagged MITF DNA-binding motif, followed by streptavidin pull-down. Streptavidin pull-down was then immunoblotted for indicated proteins. (H) MCF-7 PR cells were transfected with indicated plasmids after depletion of endogenous MITF by shRNA, followed by qPCR to examine the expression of indicated genes. (I) Cell viability was examined in MCF-7 PR cells with indicated treatments. Data represent means ± SD from three independent experiments. **, p£0.01. (J-K) Representative images (J) and growth curves (K) of MCF-7 PR xenograft tumors with indicated treatments for 3 weeks. Data are represented as means ± SEM, n = 6 mice/group. **, p£0.01.