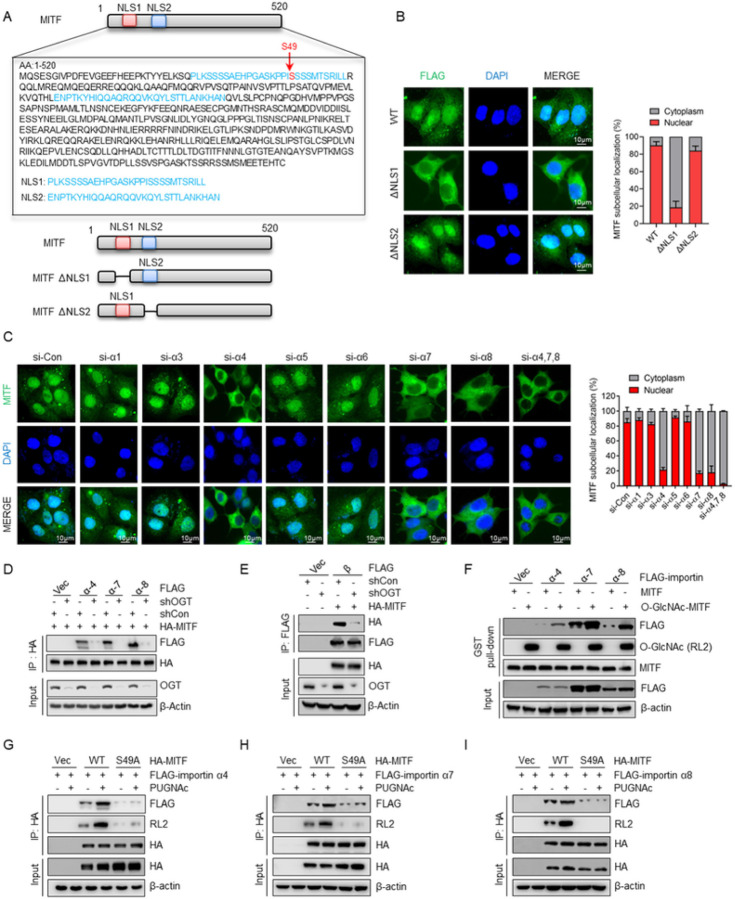
Figure 5. O-GlcNAcylation of the nuclear localization signal (NLS) promotes the interaction of MITF with importin α/β and nuclear translocation.
(A) Amino acid sequence of the MITF putative nuclear localization signals (NLSs). Locations of putative NLS1 and NLS2 were highlighted in blue, and MITF O-GlcNAcylation sites were highlighted in red. The NLS was analyzed using http://nls-mapper.iab.keio.ac.jp/. (B) Endogenous MITF was depleted in MCF-7 PR cells, followed by transfected with indicated plasmids, and MITF cellular localization was examined by immunostaining. Right, the quantification results are shown on the left. (C) MCF-7 PR cells were transfected with indicated siRNAs, and MITF cellular localization was detected by immunostaining. Right, the quantification results are shown on the left. (D) MCF-7 PR cells transfected with the indicated plasmids were harvested for co-IP. HA-IP was then resolved by SDS-PAGE and immunoblotted for indicated proteins. (E) MCF-7 PR cells transfected with the indicated plasmids were harvested for co-IP. HA-IP was then resolved by SDS-PAGE and immunoblotted for indicated proteins. (F) GST-MITF with O-GlcNAcylation was generated by in vitroglycosylation assay. MCF-7 PR cells were transfected with the indicated importin α or vector, and cell lysates were then incubated with GST-MITF with or without O-GlcNAcylation. GST pull-downs were then immunoblotted for indicated proteins. (G-I) MCF-7 PR cells transfected with the indicated plasmids were harvested for co-IP. HA-IP was then resolved by SDS-PAGE and immunoblotted for indicated proteins. The interactions of MITF with importin α4 (G), α7 (H), and α8 (I) were examined.