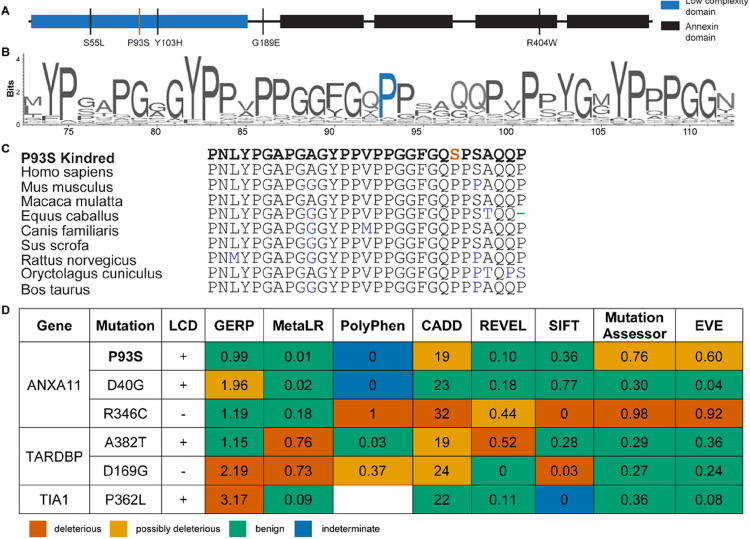
Figure 2. ANXA11 Structure.
(A) Schematic of ANXA11. Low complexity domain is shown in blue and annexin domains are shown in black. The P93S variant is identified by a red line; remaining variants/mutations are indicated by black lines. (B) and (C) ANXA11 sequence conservation. VarSite illustration of sequence conservation with the proline in position 93 indicated in blue (B) and sequence conservation of annexin A11 across vertebrate species (C) with the P93S variant indicated in red. (D) Poor performance of in silico variant prediction models for mutations in LCDs. P93S VUS with other known pathogenic mutations in common amyotrophic lateral sclerosis/frontotemporal dementia associated genes with LCDs are shown. The predictions of pathogenicity by color: predicted benign in green, moderate in orange, pathogenic in red, and indeterminate in blue. GERP score considered deleterious >2. MetaLR score ranges from 0 benign to 1 deleterious. PolyPhen score considered deleterious >0.446. CADD score considered deleterious >30. REVEL score ranges from 0 benign to 1 deleterious. SIFT score considered deleterious <0.05. Mutation Assessor score ranges from 0 benign to 1 deleterious. EVE score considered potentially deleterious >0.5 and deleterious >0.7.