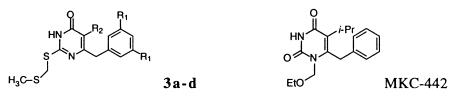TABLE 2.
Interaction scores, calculated Ki values, and biological assay data for inhibitors of HIV RT
| Compound | R1 | R2 | MSa (Å2) | BSb (%) | Lipo score | LudicKi (μM) | IC50[rRT]d (μM) | IC50[p24]d (μM) | CC50[MTA]e (μM) | Minimum SIf |
|---|---|---|---|---|---|---|---|---|---|---|
| 3a | H | Meg | 275 | 88 | 709 | 3.3 | 28.8 ± 10.0 | 3.4 ± 1.1 | >100 | 32 ± 10 |
| 3b | H | Eth | 283 | 88 | 730 | 2.0 | 12.0 ± 2.3 | 0.8 ± 0.2 | >100 | 146 ± 38 |
| 3c | H | i-Pri | 301 | 89 | 785 | 0.56 | 5.6 ± 2.4 | <0.001 ± 0.000j | >100 | 100,000 ± 0 |
| 3d | Me | i-Pr | 329 | 89 | 875 | 0.05 | 7.0 ± 2.2 | 0.016 ± 0.011 | >100 | 31,520 ± 31,310 |
| MKC442 | NAk | NA | NDl | ND | ND | ND | 0.8 ± 0.1 | 0.004 ± 0.001 | >100 | 25,830 ± 4,383 |
| Delavirdine | NA | NA | ND | ND | ND | ND | 2.3 ± 1.0 | ND | ND | ND |
| Nevirapine | NA | NA | ND | ND | ND | ND | 19.8 ± 5.5 | ND | ND | ND |
MS, molecular surface area calculated by using Connolly’s MS program (13), is defined as the boundary of the volume within any probe sphere (meant to represent a water molecule) with a given radius sharing no volume with the hard sphere atoms which make up the molecule. The values are slightly lower than those approximated by the Ludi program.
BS, the buried surface, is the percentage of the molecular surface in contact with protein calculated by Ludi relative to docked positions. Based on published crystal structures of RT complexes, our calculation shows that these values can be as low as 77% (in an RT-HEPT complex) and as high as 90% (in an RT-APA complex), but most of them, including that for RT-MKC, average around 84%.
Ludi Ki values were calculated based on the empirical score function in the Ludi program (6, 37). Ideal hydrogen bond distances and angles between compounds and protein are assumed in all cases for Ludi Ki and Ludi score calculations. In published crystal structures of RT complexes, hydrogen bond geometries are indeed close to ideal; the amide carbonyl of residue A101 on a loop demonstrates substantial flexibility, which can accommodate the best geometry for hydrogen bonding. The number of rotatable bonds (two) is used in the Ludi calculation to reflect the loss of binding energy due to freezing of internal degrees of freedom.
Mean ± standard error of three or four independent experiments (two experiments for 3a), each performed in triplicate.
Even at an inhibitor concentration of 100 μM, the viability of cells was minimally affected, with 50% cytotoxic concentration (CC50[MTA]) values of >100 μM in each of three or four independent experiments.
SI, the selectivity index, is equal to the ratio of CC50[MTA] to IC50[p24]. Since the CC50 values for the inhibitors tested were >100 μM, only the minimum selectivity index estimates could be determined for these noncytotoxic agents.
Me, methyl.
Et, ethyl.
i-Pr, isopropyl.
In each of four independent experiments, at a 1 nM concentration of 3c, the percent inhibition of p24 production was >50%, with a mean value of 67% ± 4%. In a dose-response study using picomolar and nanomolar concentrations of compound 3c, the subnanomolar IC50 was determined to be 130 pM.
NA, not applicable.
ND, not determined.