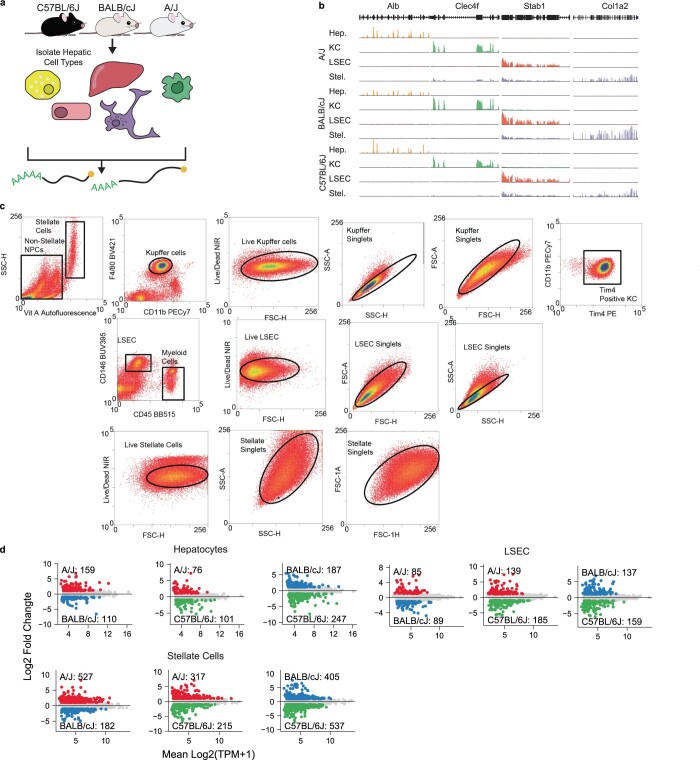
Extended Data Fig. 5. Data generation for network analysis.
a, Experimental schematic for isolation of hepatic cell types from inbred strains. b, Assessment of cell isolation purity at via RNA-seq signal at cell specific gene expression loci. Hepatocytes (yellow shades), Kupffer cells (green shades), and liver sinusoidal endothelial cells (LSEC) (red shades) were sorted with <1% contamination. Hepatic stellate cell (purple shades) RNA-seq libraries displayed minor (<10%) contamination with LSECs and Kupffer cells (seen as RNA-seq signal in Clec4f and Stab1 loci). c, FACS strategy for stellate, LSEC, and Kupffer cell isolation. d, Strain-specific transcriptional variation in hepatocytes, LSECs, and Stellate cells. N = 4 samples per subgroup.