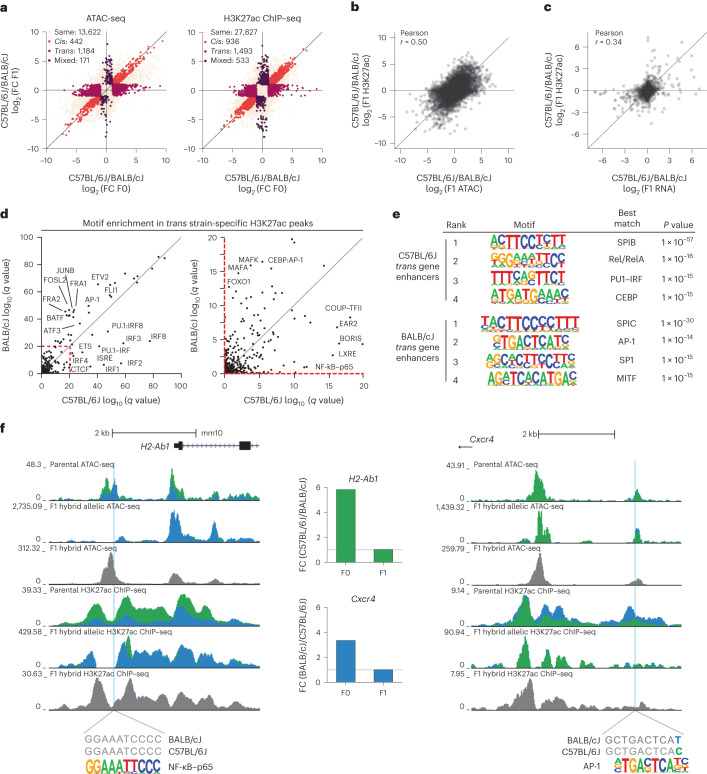
Fig. 5. Cis and trans analysis of epigenetic loci reveals upstream regulators of trans transcriptional diversity.
a, Comparison of ATAC-seq (left) and H3K27ac ChIP–seq (right) tag ratios using Kupffer cells from parental mice (x axes) and F1 hybrid mice (y axis). All IDR ATAC-seq peaks harboring a mutation were considered for analysis. Peaks with maintained ATAC-seq tag differences in both comparisons are labeled ‘cis’ and are colored red; peaks with differential ATAC-seq tags in parental cells but not in the F1 model are labeled ‘trans’ and are colored purple; peaks with similar ATAC-seq tags in parental data and allelic bias in ATAC-seq tags in F1 mice are labeled ‘mixed’ and are colored dark purple; peaks with similar ATAC-seq tags in both comparisons are labeled ‘same’ and are colored beige. b, Correlation of allelic ATAC-seq and H3K27ac ChIP–seq reads at IDR ATAC-seq peaks with at least 16 ATAC-seq and H3K27ac reads in at least one sample. c, Correlation of allelic RNA-seq reads and promoter H3K27ac ChIP–seq reads for transcripts expressed at TPM > 4 and promoters with greater than eight H3K27ac ChIP–seq reads in at least one sample. d, Enrichment score of known transcription factor motifs in trans BALB/cJ-specific enhancers (y axis) and in trans C57BL/6J H3K27ac enhancers (x axis). e, De novo motif enrichment at active enhancers (>32 H3K27ac tags) associated with C57BL/6J or BALB/cJ trans genes. f, Examples of trans-regulated epigenetic loci upstream of trans-regulated genes. A strain-invariant NF-κB motif lies within a trans-regulated C57BL/6J-specific enhancer upstream of the C57BL/6J trans gene H2-Ab1 (left). An AP-1 motif with a mutation outside the core binding sequence lies in a trans-regulated BALB/cJ-specific enhancer upstream of the BALB/cJ-specific trans gene Cxcr4 (right). Colored tracks/bars denote BALB/cJ (blue) or C57BL/6J (green) data.