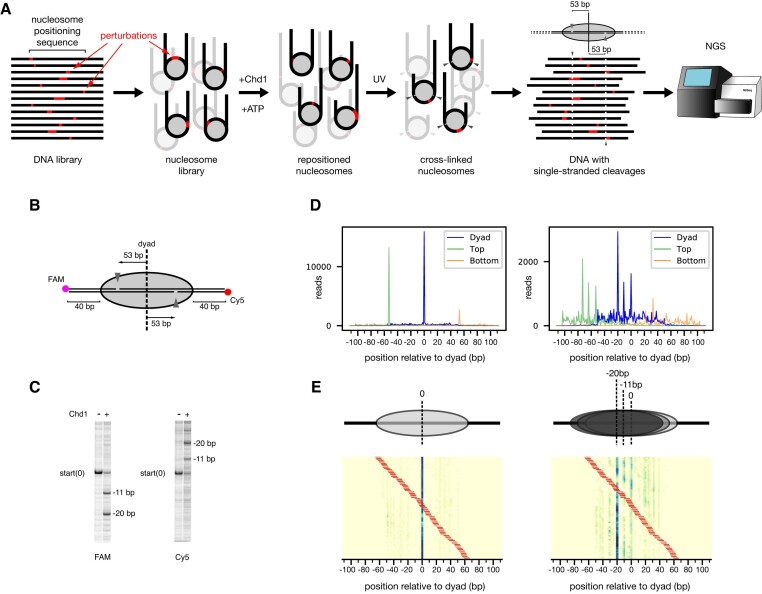
Figure 1.
(A) Schematic workflow of Slide-seq. After synthesis of a DNA library, based on a nucleosome positioning sequence, the DNA is incorporated into nucleosomes, and the nucleosomes are then repositioned by the Chd1 remodeler. To determine nucleosome positions, DNA is site-specifically photo-cross-linked to histones, which cleaves the DNA backbone 53 bp from the dyad. Next-generation sequencing these DNA fragments reveals the locations of DNA cleavages and thus the positions of DNA on the histone core for each distinct sequence. (B) Schematic of the photo-cross-linking sites on the nucleosome. When histone H2B(S53C) is labeled with azido phenacyl bromide (APB), it cross-links to only one strand of the duplex on each side of the nucleosome, 53 bp from the dyad. (C) Example of cleavage products resulting from photo-cross-linking nucleosomes, before and after sliding by Chd1. Here, the photo-crosslinked DNA cleavage was visualized on a urea denaturing gel, scanned separately for FAM (top strand) and Cy5 (bottom strand). (D) Next-generation sequencing data revealing the location of DNA cleavage sides, before (left) and after sliding (right) by Chd1. Green peaks represent cleavage sites on the top strand, orange peaks on the bottom strand, and blue peaks are calculated dyad positions. The DNA sequence for this experiment was the canonical Widom 601 DNA. (E) An example heatmap from the 601 poly(dA:dT) library with 8 bp tract length, before (left) and after sliding (right) by Chd1. Tract location is indicated by the red bars. Cartoons above the heatmaps show the most populated nucleosome positions.