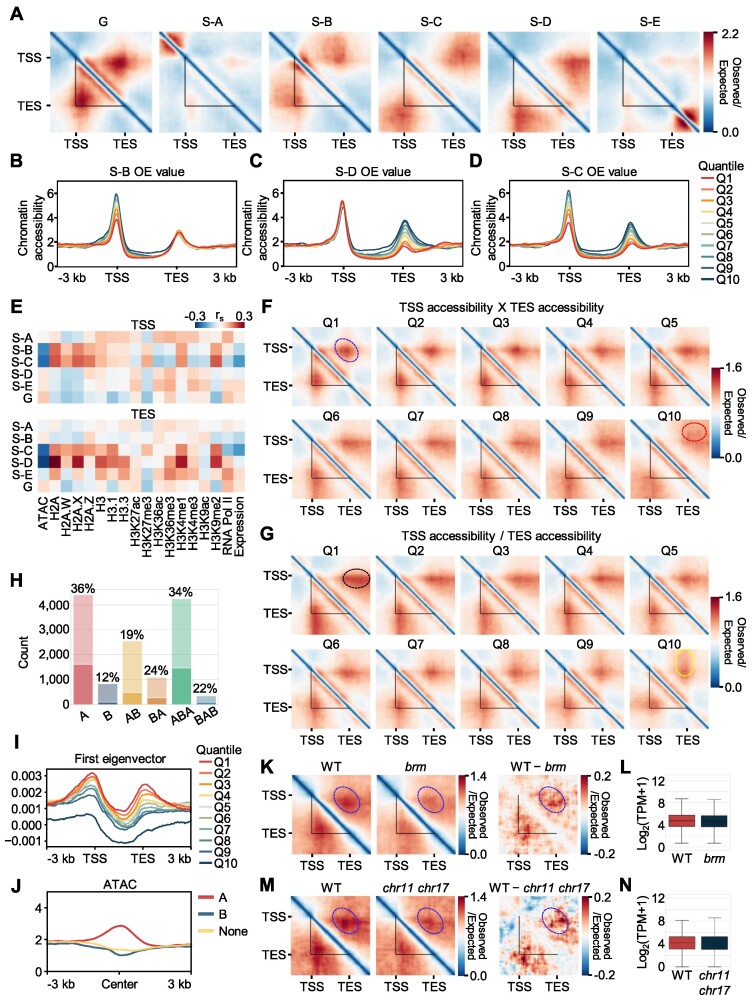
Figure 3.
Chromatin accessibility is responsible for the formation of single-gene domains. (A) Pile-up images of Hi-C contact matrices of Arabidopsis genes with the top 10% OE values in each subregion. (B–D) Metaplots of chromatin accessibility for each OE value quantile. Accessibility levels in each OE value quantile at S-B (B), S-D (C) and S-C (D) regions are shown. (E) Correlation heatmaps between the sum of chromatin contacts at each subregion and the enrichment level of chromatin features at TSS and TES. Spearman's correlation coefficients (rs) are shown. (F) Domain structures of genes clustered by TSS accessibility × TES accessibility. Genes longer than 2 kb were clustered into 10 quantiles according to the values of TSS accessibility × TES accessibility. (G) Domain structures of genes clustered by TSS to TES accessibility ratio (TSS accessibility/TES accessibility). Genes longer than 2 kb were divided into 10 quantiles according to the ratio between TSS and TES accessibility. Black circles indicate extended contacts between TSS and TES-downstream sequence, whereas yellow circles indicate extended contacts between TSS-upstream sequence and TES. (H) Number of genes with strong gene domains in each local A/B compartment defined at 5-restiction-fragment resolution. Genes within Q1–Q3 quantiles clustered by TSS accessibility × TES accessibility were annotated as strong gene domains. Boxes with light colors indicate the total number of Arabidopsis genes contained in each local A/B compartment. The ratio of the number of genes with strong gene domains over total number of genes in each local compartment is provided. (I) Metaplot of the first eigenvector of the Pearson's correlation matrix of a normalized Hi-C contact map. The average values of the first eigenvector at genic regions of genes in each quantile divided by the values of TSS accessibility × TES accessibility are shown. (J) Metaplot of chromatin accessibility within each local compartment at genic regions. Fragments were annotated as A, B or none based on their first eigenvector values. (K, M) Comparison of gene domain structures between the wild type and the chromatin remodeler mutants brm and chr11 chr17. Pile-up images of Hi-C contact matrices of target genes of BRM (longer than 2 kb, n = 1 904) (K) and CHR11 (longer than 2 kb, n = 830) (M) chromatin remodelers are shown. Left panel, wild-type Hi-C contact map; middle panel, Hi-C contact map in the chromatin remodeler mutant; right panel, difference in Hi-C contact maps between the wild type and the chromatin remodeler mutant. (L, N) Expression levels of chromatin remodeler target genes in wild type and chromatin remodeler mutants. Log2(transcripts per million [TPM] + 1) values of BRM-target genes (longer than 2 kb, n = 1 904) (L) and CHR11-target genes (longer than 2 kb, n = 830) (N) are shown. In (F), (K) and (M), the red circles indicate S-C subregion, whereas the blue circles indicate a TSS–TES contact site. In (A), (F), (G), (K) and (M), the black triangles indicate gene boundaries.