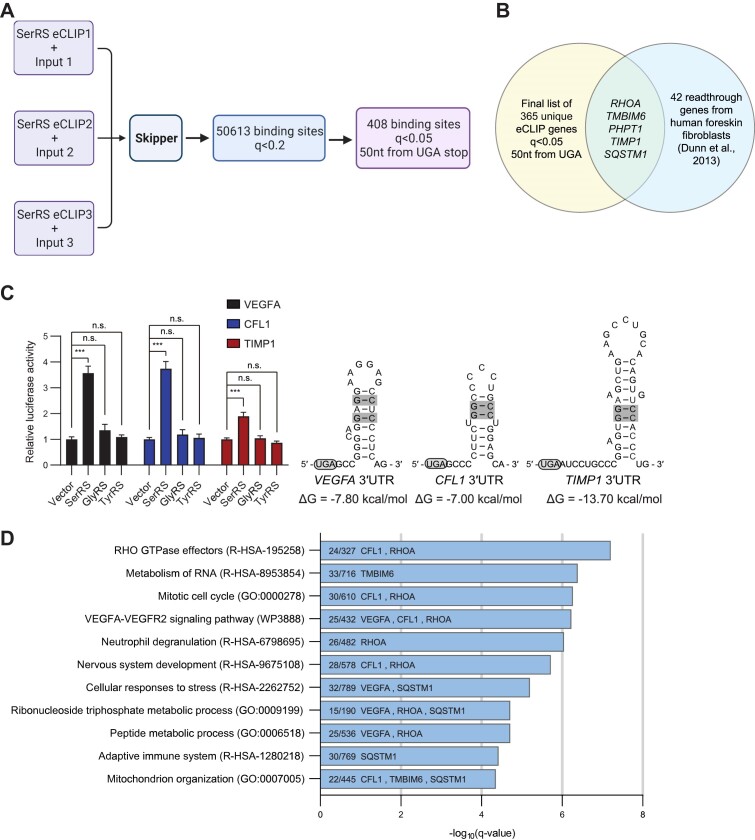
Figure 4.
eCLIP-seq identifies a set of SerRS-bound RNAs that includes other translational readthrough genes. (A) A schematic showing the eCLIP-seq analysis workflow and filtering steps to reach the final list of 408 SerRS-bound windows within 50 nucleotides of a UGA stop codon (q < 0.05). Created with BioRender.com. (B) Venn diagram showing the overlap between the final 365 unique genes found in our eCLIP set (from 408 windows) with the confirmed translational readthrough genes identified in human foreskin fibroblasts by Dunn et al. (6). (C) Luciferase reporter assays were performed for the hits CFL1 and TIMP1, using VEGFA as a positive control. All three RNAs possess a G–C base pair-containing stem-loop after the canonical UGA stop codon. (D) Gene Ontology (GO) analysis was performed on RNAs from the final list of 408 windows. The number of genes enriched in each pathway are shown along with the overlapping translational readthrough genes from Figure 4B and our experimentally confirmed translational readthrough genes in Figure 4C. (A–D) (n.s., not significant; *** P< 0.001).