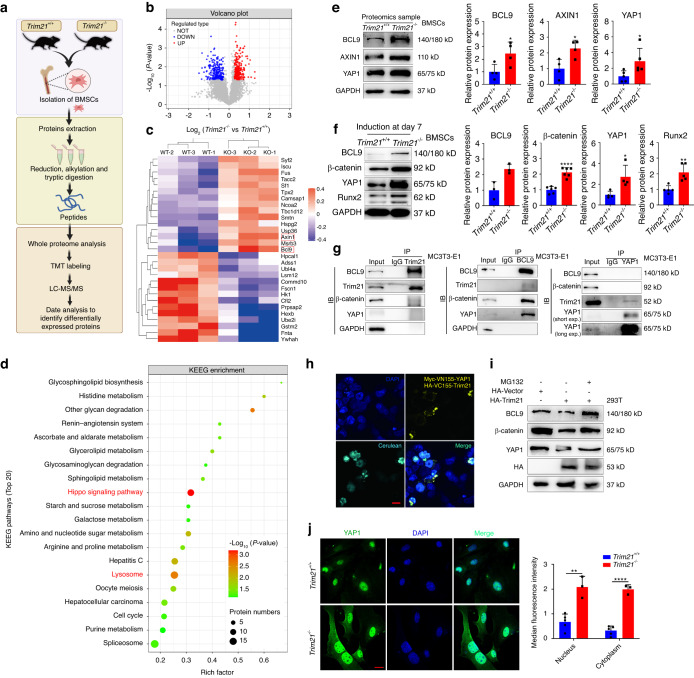
Fig. 4.
YAP1/β-catenin signaling is essential for Trim21-mediated osteogenic differentiation. a Schematic diagram showing TMT-based quantitative proteomics for the identification of differentially expressed proteins (DEPs) in bone marrow mesenchymal stem cells (BMSCs) derived from Trim21+/+ and Trim21−/− mice. b Volcano plots of DEPs in BMSCs from Trim21+/+ and Trim21−/− mice. c Heatmap analysis of DEPs in BMSCs. Three replicates of each group were included, and the top 29 DEPs are shown. d KEGG enrichment analysis of the DEPs in the BMSCs. e Representative immunoblotting analysis and quantification of BCL9, AXIN1, and YAP1 in BMSCs; proteomics sample: part of the samples subjected to proteomics analysis. f Representative immunoblotting analysis and quantification of BCL9, β-catenin, YAP1, and Runx2 protein expression in BMSCs after osteogenic induction for 7 days. g The endogenous interaction between Trim21, BCL9, β-catenin, and YAP1 was evaluated using a co-IP assay. h Protein‒protein interaction of YAP1 and Trim21 in living cells. The two BiFC plasmids encoding Myc-VN155-YAP1 and HA-VC155-Trim21 along with HA-cerulean were cotransfected into HEK293T cells for 24 h. Representative images showing transfected cells (cerulean) and the interaction between YAP1 and Trim21 (Venus). Nuclei were stained with DAPI. Scale bar: 20 μm. i Immunoblotting analysis of BCL9, β-catenin, YAP1, and HA-Trim21 protein expression in HEK293T cells treated with or without MG132. j Representative images showing the expression of YAP1+ OBs derived from Trim21+/+ and Trim21−/− mice. Scale bar: 20 μm. All bar graphs are presented as the mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.000 1; n.s. not significant by Student’s t test