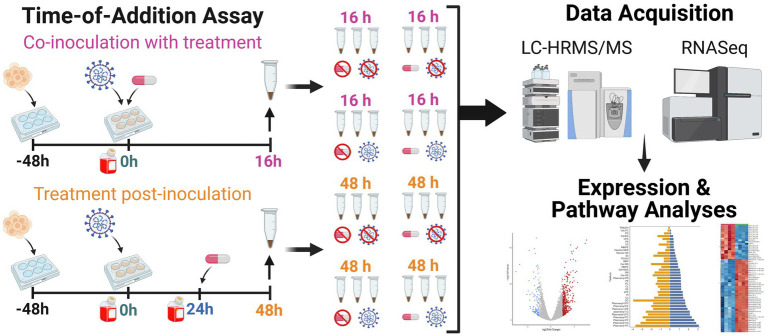
Figure 1.
Workflow for lipidomic and transcriptomic profiling of Vero E6 cells after SARS-CoV-2 infection with and without niclosamide treatment. We used a time-of-addition assay experimental design to (i) capture the lipidomic profile of SARS-CoV-2 infected Vero E6 cells, and (ii) explore the effect of niclosamide on the lipidomic profile of Vero E6 cells when added in the absence of infection, with SARS-CoV-2 virus, or at 24 h post-infection. Each replicate experimental condition (n = 3) was processed for LC-HRMS/MS or RNASeq analyses. Samples were seeded 48 h prior to the start of the experiment (t = 0 h). For all samples, media was changed at the start of the experiment (t = 0 h) and infected with virus (for infected sample groups). Sample collection is denoted by an up arrow and tube above the timeline, media changes are denoted by red bottles, addition of virus and DMSO/drug are denoted as well. Separate samples for each condition were collected for LC-HRMS/MS and RNAseq analysis, respectively. Created with BioRender.com.