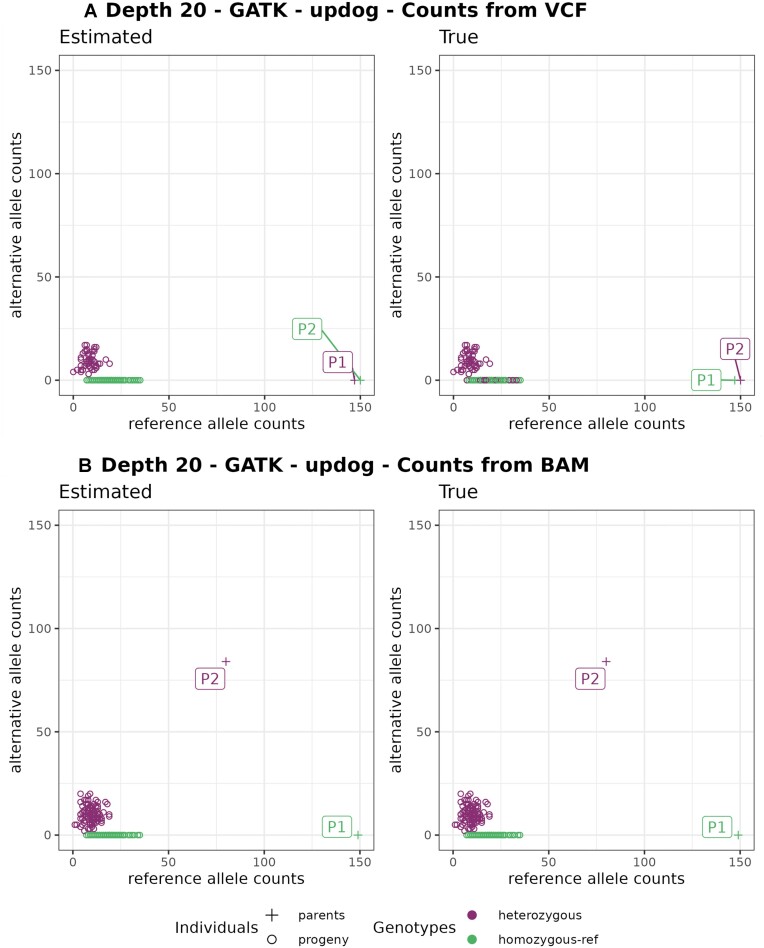
Figure 3:
Example of error (Est: homozygous | True: heterozygous and Est: heterozygous | True: homozygous) in parental genotypes leading to a wrong marker type (Est: D1.10 | True: D2.15). Estimated reference (x-axis) and alternative (y-axis) allele count. Graphics on the left have colors according to estimated genotypes and on the right to the true genotypes. (A) Counts from the GATK VCF file and (B) from the BAM file. In the VCF file outputted by GATK, the P1 genotype is missing (GT./.) because the reads did not pass the quality filters, but it reports the counts in the reference AD field (149,0). The updog software use progeny segregation (1:1) to estimate the parents, but it makes a mistake identifying which one is heterozygous. Using counts from the BAM file (B) fixes this issue despite losing the GATK quality filters that can be important in other situations.