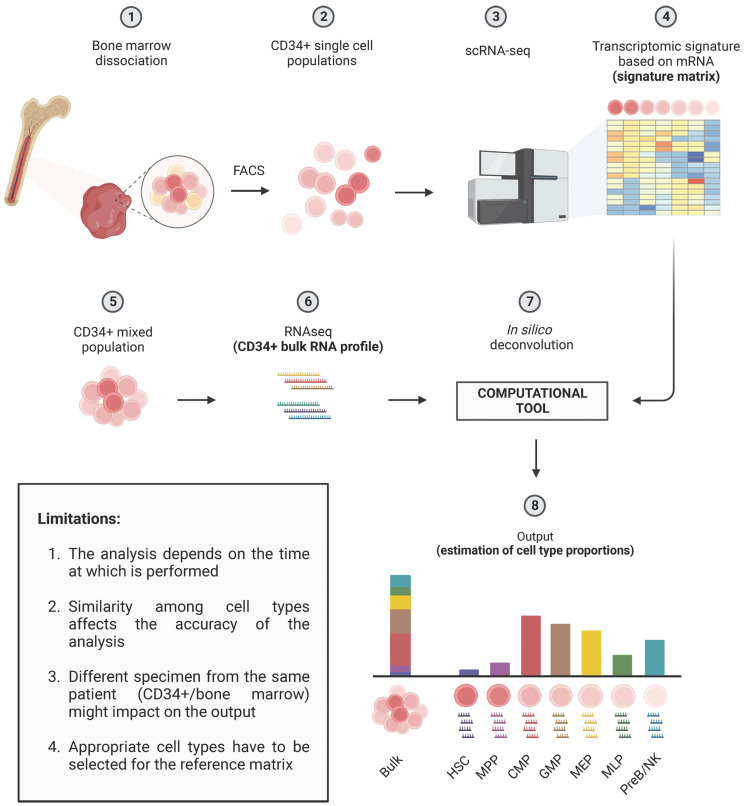
Figure 2.
Deconvolution workflow of bulk RNA-seq dataset from CD34 positive hematopoietic stem cells. Available sc-RNAseq datasets from CD34 positive cells from human bone marrows (1–3) are used to generate a signature matrix (4). In the second step, the deconvolution methods (7) require the expression profiles of CD34-positive mixed population (5–6) and the signature matrix (4) as input bulk data to estimate the abundance of cell types in each sample (8). FACS (Fluorescence-Activated Cell Sorting), HSC (Hematopoietic Stem Cell), MPP (MultiPotent Progenitor), CMP (Common Myeloid Progenitor), GMP (Granulocyte-Monocyte Progenitor), MEP (Megakaryocyte-Erythrocyte Progenitor), MLP (Multi-Lymphoid Progenitor), and PreB/NK (Pre-B cells and Natural Killer Cells).