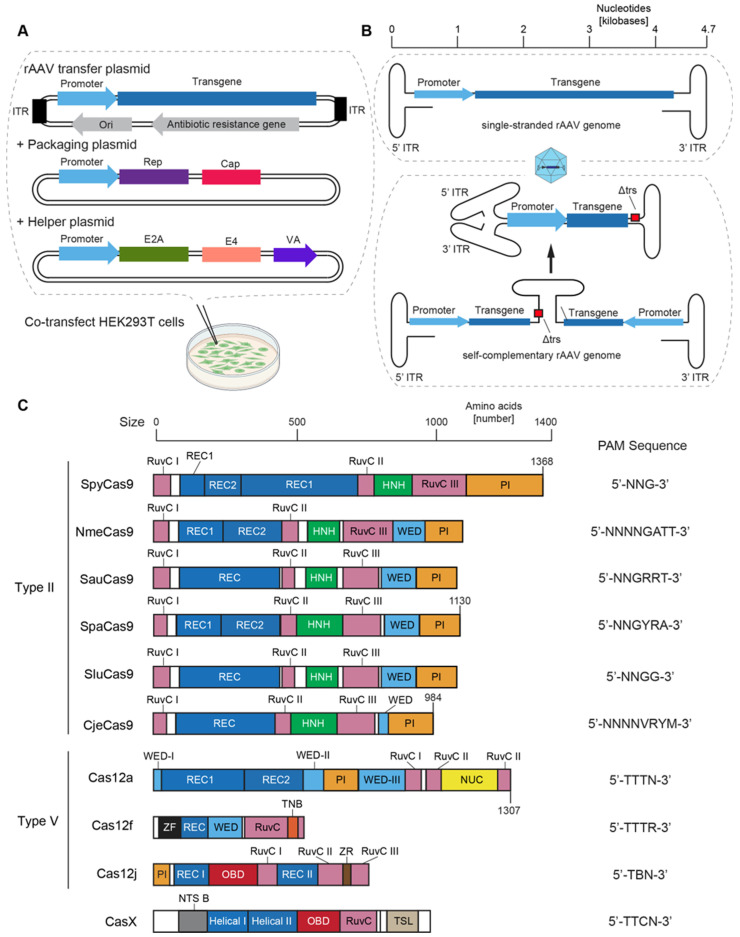
Figure 3.
Key elements and domain organization of all-in-one rAAVs. (A) Schematic of a generic rAAV transfer plasmid plus the packaging and helper plasmids that need to be co-transfected into HEK293T cells to instruct them how to assemble the rAAV vectors. (B) Cartoon depicting the difference between conventional single-strand DNA and self-complementary rAAV genomes. Note that one of the ITRs within the genome of self-complementary rAAVs needs to have its terminal resolution site deleted (Δtrs) to prevent the genome from separating into single-stranded DNA during assembly. (C) Domain organization of Cas endonucleases discussed in the text. Cas enzymes and their subdomains are displayed to scale based on their amino acids. Domains that serve the same functions are shown in identical colors. The catalytic domains RuvC and HNH are colored pink and green, respectively. Colors for other domains involved in catalysis are as follows: Nuc, yellow; TNB, brown; TSL, beige; ZF, black; ZR, brown. Nucleic acid-interacting domains are shown in shades of blue (Helical I, Helical II, REC I, REC2, REC III, WED). Remaining domains are as follows: NTSD, grey; OBD, red; and PI, orange. Key to Cas domain acronyms: NTSB, non-target strand binding domain; PI, PAM interacting domain; REC, recognition domain; TNB, target nucleic acid binding domain; TSL, target strand loading domain; WED, wedge domain; ZF, zinc finger domain; ZR, zinc ribbon domain. PAM sequences are shown in single letter nucleotide code. Incompletely specified bases follow the standard nomenclature, i.e., Y, cytosine or thymine; B, cytosine, guanine or thymine; M, adenine or cytosine; R, adenine, or guanine; N, guanine, adenine, thymine or cytosine. (C) Domain organization of all-in-one rAAVs discussed in the text that have been employed for in vivo gene editing therapies. The figure was created with Biorender.