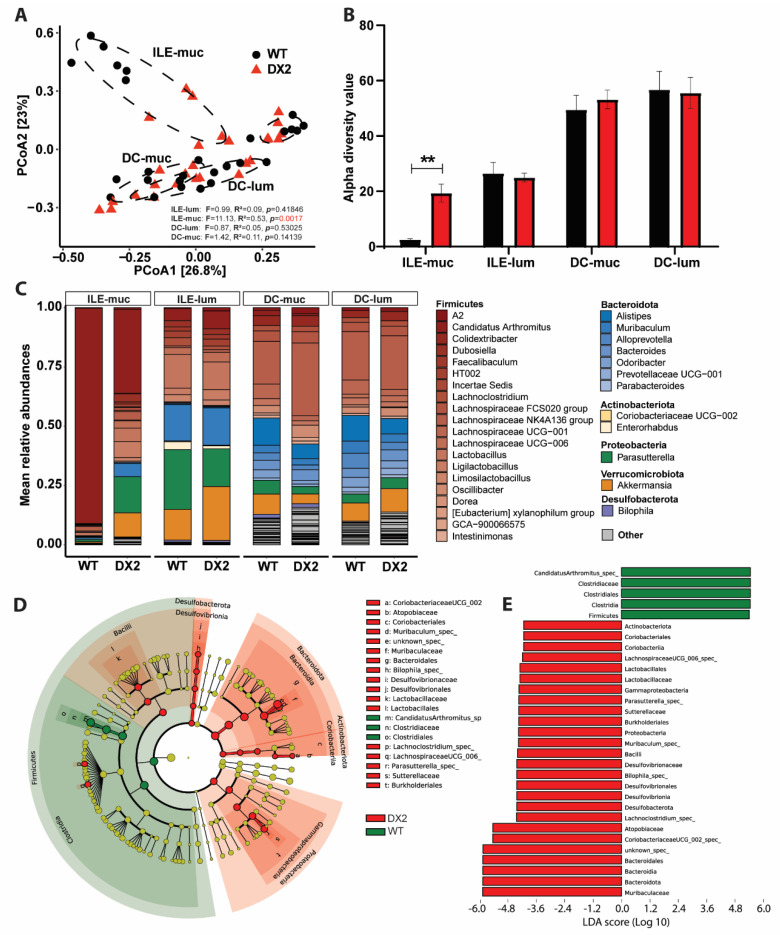
Figure 2.
Ablation of epithelial DUOX2 shapes the mucosal microbiome. Paired mucosal and luminal samples from the small intestine and colon of WT and Duox2∆IEC mice underwent 16S rRNA microbiome analysis. (A) Principal coordinate analysis revealed the different composition of the mucosal but not in the luminal microbiome in WT and Duox2∆IEC mice (WT: ILE−muc n = 6, ILE−lum n = 6, DC−muc n = 6, DC−lum n = 9. DX2: ILE−muc n = 6, ILE−lum n = 6, DC−muc n = 7, DC−lum n = 8). (B) Alpha diversity (within sample diversity) using the Inverse Simpson metric. (C) Taxonomic overview on species level color−shaded by phylum. (D,E) Linear discriminant analysis (LDA) effect size (Lefse) of the ileum mucosa microbiome. (D) The cladogram depicts the phylogenetic distribution of differential taxa. (E) Differential taxa ranked by LDA. ILE = ileum, DC = distal colon. muc = mucosal, lum = lumen. ** p < 0.01.