Figure 1.
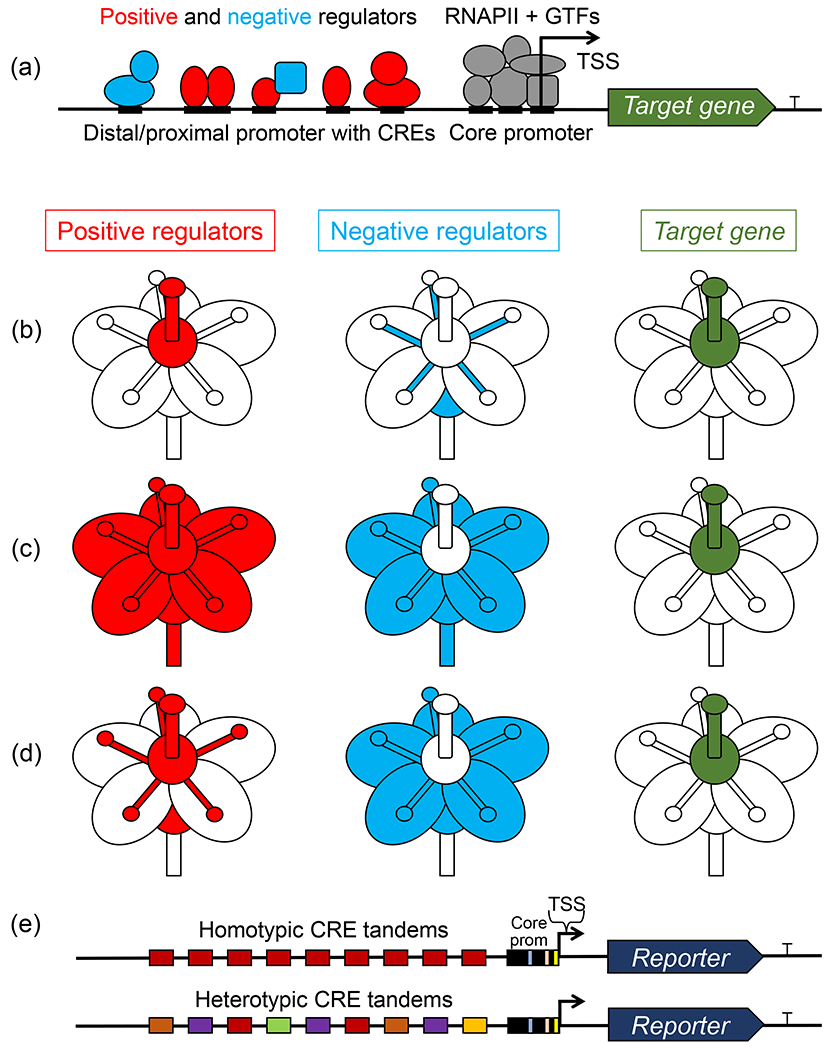
Transcriptional regulation underlying tissue specificity of gene expression. (a) Native promoters harbor multiple cis-regulatory elements (CREs) that bind a combination of positive (red) and negative (cyan) regulators (transcription factors, co-factors, and epigenetic effectors) that, respectively, assist and interfere with the RNA polymerase II (RNAPII) and general TF (GTF) (gray) recruitment to the core promoter. Arrow marks the transcription start site (TSS). (b, c, d) Different expression patterns of positive (red) and negative (cyan) regulators can result in restricted domains of target gene expression (green) if negative regulators negate the effects of positive regulators. (e) Typical synthetic reporter constructs harboring tandems of identical or divergent CREs placed upstream of a well-characterized core promoter, such as (−46)35S, driving a reporter gene, such as GFP, Luciferase, or GUS.
