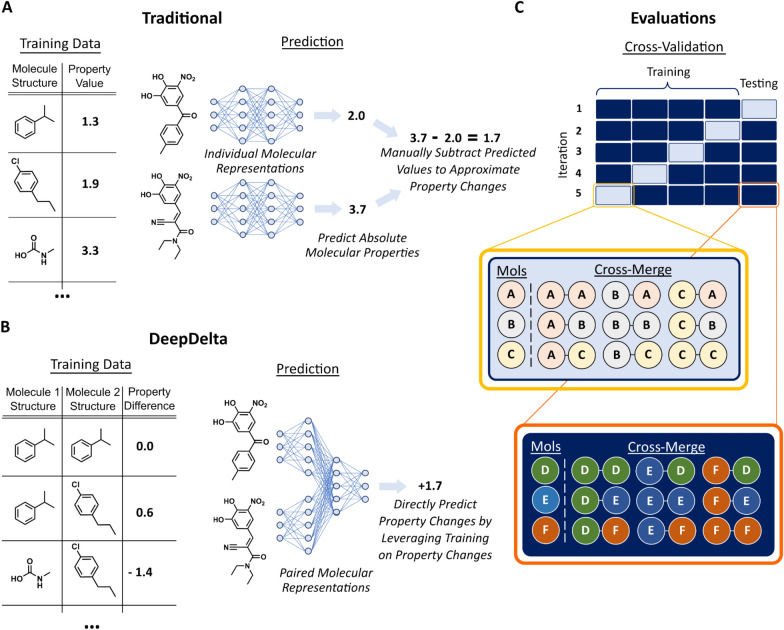
Fig. 1.
Traditional and pairwise architectures. A Traditional molecular machine learning models take singular molecular inputs and predict absolute properties of molecules. Predicted property differences can be calculated by subtracting predicted values for two molecules. B Pairwise models train on differences in properties from pairs of molecules to directly predict property changes of molecular derivatizations. C Molecules are cross-merged to create pairs only after cross-validation splits to prevent the risk of data leakage during model evaluation. Therefore, every molecule in the dataset can only occur in pairs in the training or testing data, but not both