Figure 4.
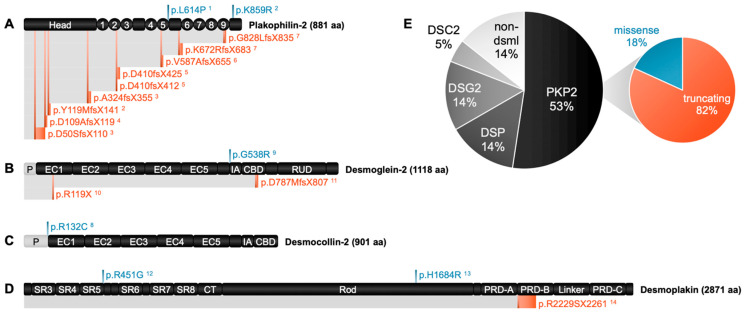
dACM-associated mutations studied in hiPSC-CMs to date. Primary structure schematics [82] for (A) plakophilin-2, (B) desmoglein-2, (C) desmocollin-2, and (D) desmoplakin are shown with all known variants studied in hiPSC-CMs mapped to their respective protein in p-dot (p.) nomenclature (superscripts denote references to primary source—see end of caption). Note that the “P” domain in light gray at the N-terminus of desmocollin-2 and desmoglein-2 represents amino acids which are part of the preproprotein and are eventually cleaved off to yield a mature protein. Missense variants are denoted in blue, and truncating variants are denoted in red. In this diagram, a line branching from a protein indicates the site where a specific mutation causes an amino acid switch, and a red band running parallel to the protein of interest from the switch site represents how far downstream the protein is translated before a premature stop codon is reached. (E) Proportion of ACM studies in hiPSC-CMs attributable to mutations of specific genes: PKP2 (plakophilin-2), DSP (desmoplakin), DSG2 (desmoglein-2), DSC2 (desmocollin-2), and non-desmosomal genes. To the best of our knowledge, mutations in JUP (plakoglobin) have not yet been studied in hiPSC-CMs. In contrast, most ACM reports in hiPSC-CMs investigate PKP2 variants, a majority of which are truncating variants over missense variants. Superscripts for references: 1 [83]; 2 [84]; 3 [58]; 4 [85]; 5 [86]; 6 [87]; 7 [8]; 8 [88]; 9 [89]; 10 [90]; 11 [59]; 12 [91]; 13 [92]; and 14 [93].