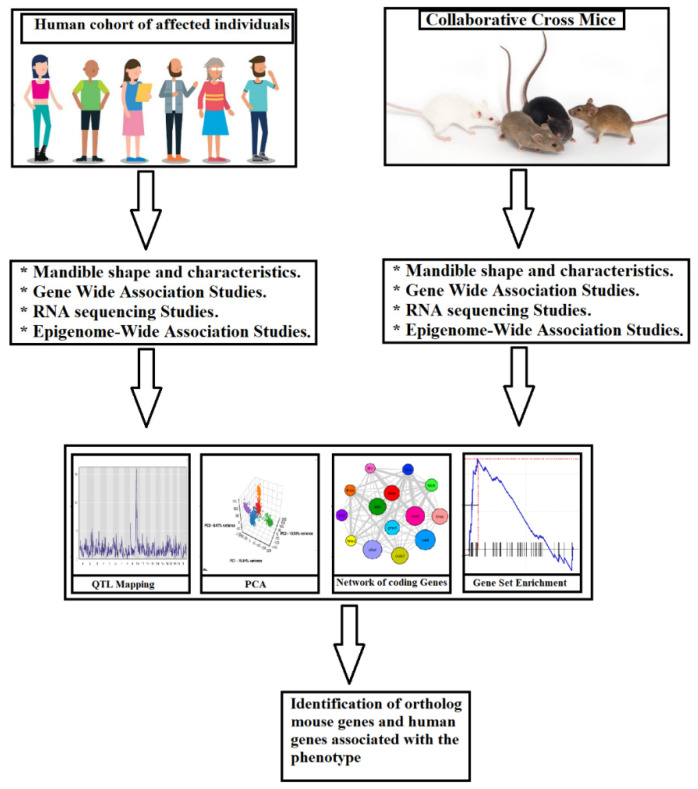
Figure 15.
Workflow for creating system genetic databases from the extremely heterogeneous CC population, whose propensity to develop deep bite malocclusion may vary greatly. To check for deep bite malocclusion, mice are examined. Then, different associations between malocclusion and deep bite characteristics are analyzed using a combination of cellular, molecular, and clinical trait data. The regulatory genomic areas are implicated in phenotypic variation in both in vitro and in vivo monitored traits and can be identified using QTL mapping by merging SNP genotype data from each CC lineage. Finding susceptibility genes linked to the emergence of deep bite in humans may be possible by combining data with later candidate gene association studies in humans.