Abstract
Mucosal melanoma (MM), an aggressive rare subtype of melanoma, is distinct from cutaneous melanoma and has poor prognoses. We addressed the lack of cell models for MM by establishing 30 organoids of human oral MM (OMM), which retained major histopathological and functional features of parental tumors. Organoid groups derived from chronologically or intratumorally distinct lesions within the same individual displayed heterogeneous genetics, expression profiles, and drug responses, indicating rapid tumor evolution and poor clinical response. Furthermore, transcriptome analysis revealed receptor tyrosine kinases (RTKs) signaling, particularly NGFR, a nerve growth factor receptor, was significantly up-regulated in OMMs and organoids from patients resistant to anti–programmed cell death protein 1 (anti–PD-1) therapy. Combining anti–PD-1 with anlotinib (a phase 2 multitarget RTK inhibitor for OMM) or NGFR knockdown enhanced the effective activity of immune cells in organoid–immune cell coculture systems. Together, our study suggested that OMM organoids serve as faithful models for exploring tumor evolution and immunotherapy combination strategies.
Patient-derived organoids model tumor heterogeneity of human mucosal melanoma and explore immunotherapy combination strategies.
INTRODUCTION
Mucosal melanoma (MM) is an aggressive rare subtype of melanoma that occurs more frequently among East Asians than Caucasians (1). It generally originates from melanocytes in mucosal membranes, with approximately 40% of cases occurring in the oral cavity. MM differs substantially from cutaneous melanoma (CM) in terms of poorer prognosis, unknown etiology, and distinct genetic variations (2). The 5-year survival rate for MM is only about 20% (3). Moreover, MMs exhibit a low–point mutation burden but a high number of structural variants compared to CMs, including amplification of oncogenes, such as AGAP2, CDK4, MDM2, and TERT (4, 5). Frequently mutated genes in CMs, such as BRAF, NRAS, and NF1, are less common in MMs, whereas somatic mutations of the c-KIT and POM121 have been reported as relatively frequent in MMs (4, 6).
There are now no approved therapies specifically targeting metastatic MM following surgical resection. The medical advice for MM is primarily based on CM treatment approaches, but the reported benefits are modest (7). For instance, immune checkpoint blockade therapy, which functions by blocking the programmed cell death protein 1/programmed death ligand 1 pathway (anti–PD-1/PD-L1), has been recommended for advanced CM. By contrast, its therapeutic effect in MM is poorer (overall response rate: CM, 40.9%; MM, 20.3%) (8), potentially due to genetic alterations in cyclin-dependent kinase 4 (CDK4) pathway components, lower expression of PD-L1, and imbalanced microecology in MMs (9, 10). Consequently, there is an urgent need for previously unidentified therapeutics or therapeutic combinations involving anti–PD-1/PD-L1 for patients with MM (11). However, the lack of public databases and cell models specific to MMs has hindered the development and validation of potential clinical options. Most of the available cell models for melanoma are CM cell lines, which do not accurately represent the genetics, biological phenotypes, or drug responses of MMs, considering the known distinctions between CMs and MMs (12, 13). Therefore, it is of paramount importance to generate MM-specific cell models, particularly those that faithfully recapitulate the parental tumor characteristics.
Three-dimensional cancer organoids offer considerable advantages over cell line models in preserving the physical architectures of tumor tissues and modeling functional interactions within the tumor microenvironment (TME) (14, 15). Given these advantages, patient-derived epithelial cancer organoids have been successfully generated and widely used to model parental tumors and investigate drug responses in various cancer types, including neuroendocrine neoplasms, cervical, pancreatic, and rectal cancers (16–19). In addition, autologous peripheral blood mononuclear cells (PBMCs) or tumor-infiltrating lymphocytes (TILs) have been used in vitro to facilitate tumor recognition and high cytotoxic activity through coculture with certain tumor organoids, as demonstrated in breast and colorectal cancer organoids (20–22). These coculture systems, which model tumor–immune cell interactions, are essential and convenient for preclinical evaluation of anti–PD-1/PD-L1 or related immunotherapy combination strategies in vitro (23). However, there is a paucity of melanoma organoid models reported or well characterized, particularly for the rare oral MM (OMM) (24).
In this study, we present the generation and analysis of 30 patient-derived OMM organoids. These organoids faithfully retained the histological features and molecular diversity of their parental tumors. With organoid groups derived from chronologically or intratumorally distinct lesions of the same individual, we identified variations in gene expression profiles, genomic alterations, and drug responses during OMM evolution. Furthermore, we demonstrated the utility of these OMM organoids as a model system for investigating drug responses and exploring treatment combinations with anti–PD-1 therapy to overcome cellular resistance. These organoids serve as a valuable cell model repository for this rare tumor type and offer opportunities for drug testing and advancements in personalized medicine approaches for OMMs.
RESULTS
A library of OMM organoids
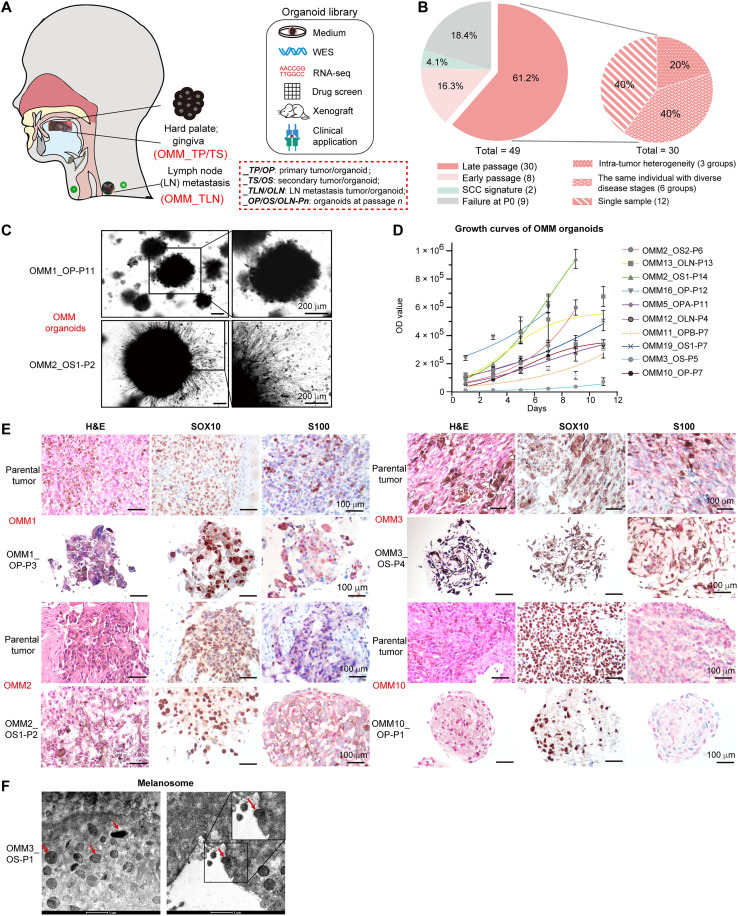
To establish models for human OMM, we obtained surgically resected tumor specimens from 41 in situ (the hard palate or gingiva of the oral cavity) and 8 lymphatic metastatic tumors of patients with OMM (Fig. 1A). OMM tissues originated from melanocytes, the neural crest–derived cells, and exhibited diffuse patterns with poor tissue structure and morphology, which differed from epithelial cells with well-organized structures (25). More than 90% of the specimens that we obtained (except OMM5_TP was amelanotic; “_TP” denoted “primary tumor”) were rich in melanin particles and lacked intercellular tight junctions in the epithelial cells (fig. S1A). Considering the above features, to optimize the processing of OMM tissues into organoids, we made small adjustments to the standard epithelial organoid processing procedures (26), including increased washing steps to remove some melanins for clear observation and shorter mild enzyme digestion for OMM tissues without tight junctions. These modifications resulted in an improved success rate of organoid establishment and serial passaging, increasing from 54 to 64% (tables S1 and S2).
Fig. 1. Establishment of patient-derived oral mucosal melanoma organoids.
(A) Overview of experimental plan. (B) Summary of the current status of cultivated OMM organoids and the heterogeneous characteristics of organoids at late passages. (C) Bright-field images of OMM organoids. Scale bars, 200 μm. (D) Representative growth curves of established organoids. OD values were acquired with CellTiter-Glo 3D cell viability assay (n = 4, data presented as the means ± SD). OD, optical density. (E) Hematoxylin and eosin (H&E) and immunohistochemistry (IHC) staining of melanoma cell marker SOX10, and S100 (red) in paraffin-embedded OMM organoids and the corresponding tissues. Scale bars, 100 μm. (F) Representative transmission electron microscopy (TEM) images showing ultrastructures of OMM organoids. Arrows, cytoplasmic location of melanosomes. Scale bars, 1 μm. For (C) to (F), “-Pn” denotes “-Passage n.”
Using this optimized approach, we successfully established 30 OMM organoids from 21 patients, with ages ranging from 28 to 73 years (clinical data in table S3, “OMM_O” denoted “OMM organoids”) (Fig. 1B). Among these organoids, 20% were derived from lymphatic metastasis, while the remaining 80% were from in situ tumors (fig. S1B). Notably, 12 organoids were derived from post–drug-treatment tissues, while 18 organoids were from pre–drug treatment (fig. S1C). Because OMM progressed rapidly, we were able to obtain specimens from chronologically distinct lesions of the same patient within a span of 2 to 6 months. In total, we generated six groups of organoids representing two disease progression statuses per individual, providing valuable resources for studying tumor evolution and comparing drug responses (Fig. 1B and fig. S1, D to H). Moreover, we generated three groups of organoids from two different regions within each large tumor lesion, allowing us to explore intratumor heterogeneity (Fig. 1B and fig. S1, I and J).
One prominent feature of the established OMM organoids was their completely black appearance in bright-field images, exhibiting diversity in growth rate and morphology (Fig. 1, C and D, and fig. S2A). On the basis of observable features at the edges of the organoids, we roughly categorized them into two groups: one showing epithelial-like spheroidal structures with a relatively smooth margin (“epithelial-like”), and the other showing invasive, loosened structures with neuronal dendrite-like branches extending from the cell bodies (“invasive-like”) (fig. S2B). These morphological differences between the epithelial-like and invasive-like organoids supported the polymorphic nature of OMMs (25) and were associated with differential molecular features, including distinct cell-cell adhesion levels and metabolic requirements (fig. S2C). However, no significant differences were observed in the recurrence and metastasis frequencies of their parental tumors (fig. S2D).
Hematoxylin and eosin (H&E) staining demonstrated strong concordance between the histopathological features of OMM organoids and their parental tumors (Fig. 1E), including cytologic pleomorphism with the cytoplasmic distribution of melanin, the latter of which was confirmed with transmission electron microscopy (TEM) images (Fig. 1F). The organoids were also positive for melanoma markers (S100, SOX10, MELANA, and GP100), as well as melanosome markers (TYRP1 and LAMP2) and proliferation markers (Ki67 and p-mTOR) (27, 28) (Fig. 1E and fig. S2, E to H). Karyotyping revealed aberrantly higher chromosome numbers in OMM2_OS2 and OMM13_OLN (“_OS” denoted “secondary organoid” and “_OLN” denoted “lymph node metastasis organoid”), suggesting chromosomal instability preserved in the organoids (fig. S2, I and J) (29). These findings collectively confirmed the melanocytic origin of the generated organoids, which exhibited rapid proliferation and retained the histopathological characteristics of the original OMMs (30).
Organoids recapitulating the major genetic variations of their parental OMMs
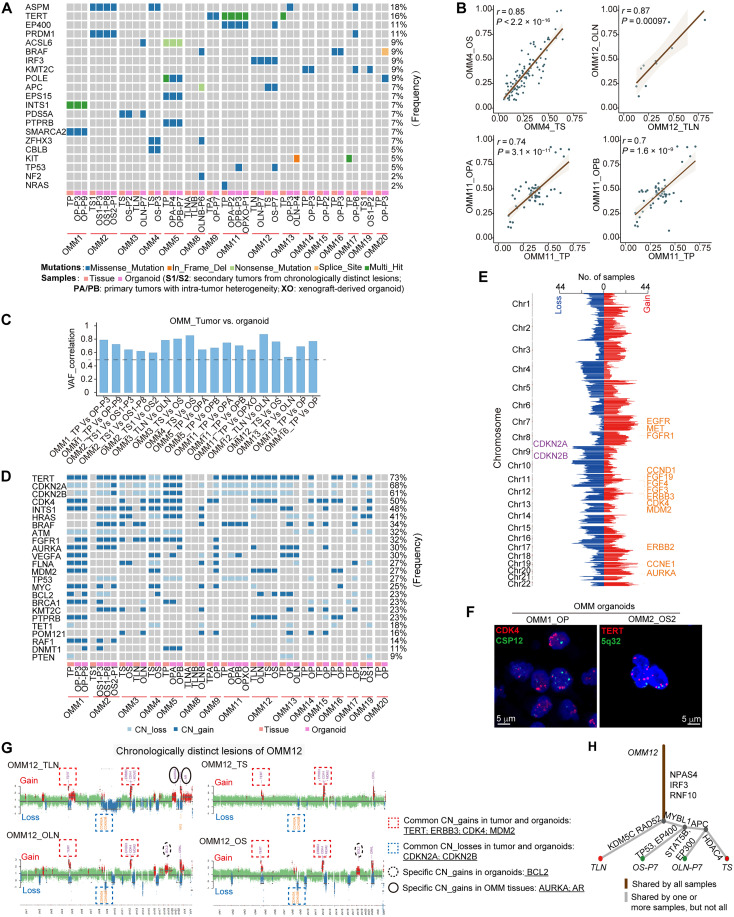
To assess somatic mutations and DNA copy number variations (CNVs) in the organoids and determine their genetic representativity for the corresponding tumors, we performed whole-exon sequencing (WES). Major parental mutations, such as ASPM (18%), TERT (16%), and EP400 (11%), were retained in the OMM organoids (Fig. 2A). Linear regression analysis of the variant allele frequency (VAF) distributions demonstrated that OMM organoids retained at least 60% of the variants detected in their parental tumors, a significantly higher rate compared to unmatched pairs (Fig. 2, B and C, and fig. S3A). In cases of OMM8 and OMM9, whose parental OMM tissues had lower VAFs [correlation coefficient (r) < 0.2] and hardly any common driver mutations, little mutational similarities were observed between the two regions of the parental OMMs, indicating higher intratumor heterogeneity in these tumors (fig. S3B). Pair-wise comparisons between OMMs and their corresponding organoids highlighted strong concordance for CNVs at a genome-wide scale (fig. S3C). OMM organoids retained various chromosomal changes, including gains of 5p15 (TERT), 7q11 (POM121), 7q34 (BRAF), and 12q13-15 (CDK4 and MDM2) chromosomal regions, as well as losses of the 9p21 (CDKN2A/B) and 17q15 (TP53) chromosomal regions, consistent with previous data (Fig. 2, D and E) (4). Furthermore, fluorescence in situ hybridization (FISH) validated the CNV amplifications of CDK4 and TERT in the corresponding OMM organoids (Fig. 2F). These results supported the overall genetic representativity of the OMM organoids to their parental tumors.
Fig. 2. OMM organoid lines recapitulated major genetic alterations of their parental tumors.
(A) Somatic mutations identified through whole-exon sequencing (WES) in OMM organoids and their parental tumors are summarized. (B) Scatter plots display the correlation between the DNA variant allele frequency (VAF) of OMM organoids and their parental tumors. The figures show the linear regression line (brown) and the corresponding r and P values. (C) Waterfall plots demonstrate the VAF correlation values between OMM organoids and their parental tumors, highlighting significant correlations (P < 0.05). (D) Summary of DNA copy number alterations identified through WES in OMM organoids and their parental tumors. (E) The number of samples with gains (log2Ratio > 0.1) or losses (log2Ratio < −0.1) in corresponding chromosome regions is presented. Chr, chromosome. (F) Representative fluorescence in situ hybridization (FISH) analysis of CDK4 (left) and TERT (right) amplification in OMM organoids. Scale bars, 5 μm. (G) Scatterplots illustrating genome-wide copy number alterations of OMM tissue-organoid pairs in OMM12, with DNA copy number gains (red) and losses (blue) conserved in derived organoid lines. CN_gains, copy number gains; CN_losses, copy number losses. (H) Phylogenetic trees based on somatic mutation variations in OMM12.
To evaluate tumor evolution- or heterogeneity-related genomic alterations in this rapidly progressing tumor, we focused on sample groups from distinct lesions of the same patients, either chronologically (OMM12) or intratumorally (OMM11), as well as organoids obtained during serial passaging (OMM1), with CNV and phylogenetic analyses of WES data (Fig. 2, G and H, and fig. S3, D to G). The most common CNVs observed in OMM tissues and generated organoids were consistent, highlighting the amplification of TERT, ERBB3, CDK4, MDM2, or BRAF, as well as the loss of CDKN2A and CDKN2B in OMM1, OMM11, and OMM12. These findings indicated potential driving forces behind the tumors (Fig. 2G and fig. S3, D and F). Notably, we observed high tumor purity in organoid models of OMM1, OMM11, and OMM12, particularly in late-passage organoids. However, certain CNVs were exclusively detected in the organoids during OMM organoid generation or serial passaging, such as BCL2 in OMM12, CCND3 and VEGFA in OMM11, and FGF19 and CCND1 in OMM1 (Fig. 2G and fig. S3, D, F, and H). Furthermore, some CNVs might be associated with tumor evolution or heterogeneity and were identified only in particular OMM tissues, for instance, AURKA and AR in OMM12_TLN but not in OMM12_TS (Fig. 2G). In addition, the phylogenetic trees of pairwise OMM samples predicted mutational signatures. In OMM12, tumor-evolution-associated mutations (largest branches), including KDM5C, HDAC4, and APC; organoid-specific mutations such as EP400 and EP300; and common mutations (the trunk) such as IRF3 and RNF10 were discovered (Fig. 2H). Understanding tumor evolution- and heterogeneity-associated genomic alterations provides insights into their potential etiology and poor responses to targeted drugs and informs better treatment strategies in the future. Notably, exposure to smoking and spontaneous deamination of 5-methylcytosine mutational signatures were involved in the tumor-associated alterations observed in these OMMs, further supporting their role as underlying factors for this rare tumor (fig. S3, I and J).
Transcriptomic characterization of OMM organoids
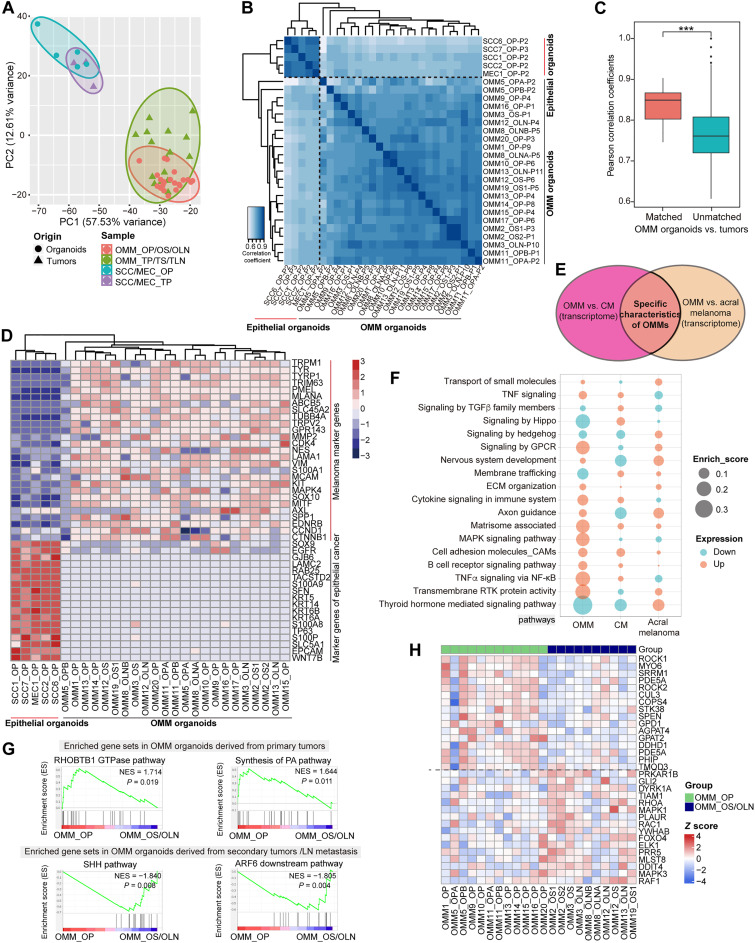
To gain insights into the gene expression profiles of OMM organoids, we performed RNA sequencing (RNA-seq) on the organoids and their parental tumors and compared them to oral epithelial cancers, namely, oral squamous cell carcinoma (SCC) and mucoepidermoid carcinoma (MEC) (RNA-seq for SCC/MEC organoids and their matched tumors, see Materials and Methods). Principal components analysis (PCA) revealed that OMM organoids clustered closely with OMM tissues, which were distinct from the sequenced SCC/MECs (Fig. 3A). Spearman correlation analysis of the transcriptome data confirmed a higher correlation between OMM organoids and OMMs (inter-SCC/MECs, r > 0.85; inter-OMMs, most r > 0.8; and OMM-SCC/MEC, r < 0.8; Fig. 3B). Moreover, Pearson correlation analysis demonstrated significant correlations between matched organoids and their parental tumors (P < 0.0001; Fig. 3C). These findings collectively suggested that the OMM organoids maintained the gene expression profiles of OMMs. In addition, compared to SCC/MECs, OMM organoids exhibited enrichment in melanoma-associated gene ontology (GO) terms, such as neurogenesis, pigmentation, and cell migration (fig. S4, A and B). OMM organoids also displayed high expression of identical melanoma marker genes while down-regulating typical markers for epithelial cancers, indicating the preservation of typical melanoma markers expression during in vitro organoid culture (Fig. 3D).
Fig. 3. Transcriptome analysis of OMM organoid lines and their parental tumor tissues.
(A) Principal components analysis (PCA) demonstrates the transcriptome data of OMM organoids, corresponding OMM tissues, SCC/MEC organoids, and their parental tumors. The figure shows the first two principal components (PC1 and PC2), where each data point represents one sample (circle for organoid and triangle for tissue). (B) Correlation heatmap between OMM organoid lines and SCC/MEC organoid lines, highlighting closer grouping of OMM organoids. (C) Pearson correlation coefficients between OMM organoids and their parental tumor tissues using 1308 highly correlated genes. “Matched” indicates matched OMM organoids and tissues pairs, while “Unmatched” refers to unmatched organoids and OMM tissues pairs. ***P = 2.63 × 10−5 by Wilcoxon rank sum test. (D) Heatmap depicting the expression of identical marker genes of melanoma and epithelial cancers in corresponding organoids. Blue indicates low expression, while red indicates high expression. (E) Venn diagram of DEGs gained after comparing OMMs to CM and acral melanoma. The intersection area represents specific characteristics for OMMs, while specific characteristics for CM and acral melanoma are similarly derived. (F) Bubble plots summarize the enriched signaling pathways in three types of melanomas: OMMs, CMs, and acral melanomas. Orange indicates positive enrichment in the melanoma subtype, while blue indicates negative enrichment. Larger bubbles indicate higher enrichment. TGFβ, transforming growth factor–β; GPCR, G protein–coupled receptor; NF-κB, nuclear factor κB. (G) Gene set enrichment analysis (GSEA) results show the enriched gene sets in OMM organoids derived from primary tumors (“OMM_OP,” top column) and in OMM organoids derived from recurrent and metastatic tumors (“OMM_OS/OLN,” bottom column). The top two pathways with higher normalized enrichment score (NES) are listed. P < 0.05 indicates significance. GTPase, guanosine triphosphatase; LN, lymph node. (H) Heatmap showing expression levels for representative genes belonging to these enriched pathways.
Previous studies have highlighted the distinct genomic landscape of MMs and acral melanoma compared to CMs (6). To investigate whether there are distinct expression profiles in MMs, we integrated our transcriptome data with published RNA-seq data of CMs and acral melanoma (see Materials and Methods) (31). Given the relatively small number of samples for comparison, we performed multiple comparisons of canonical pathways between any two of the three melanoma subtypes. The results revealed that gene clusters related to cytokines, tumor necrosis factor–α (TNFα), B cell receptor, and mitogen-activated protein kinase (MAPK) signaling were relatively up-regulated in OMMs (enrichment score, >0.1; Fig. 3, E and F). In contrast, membrane trafficking, hippo, and hormone signaling pathways were up-regulated in CMs or acral melanoma but down-regulated in MMs (Fig. 3F). Although OMMs have been reported to respond more poorly to immunotherapy compared to CMs in clinical settings, we found that OMMs were enriched in immune-related pathways (Fig. 3F) (8). Using Cibersort, we deconvoluted potential immune cell fractions in OMM tissues and CMs from a cohort in The Cancer Genome Atlas (TCGA) database [downloaded from Genomic Data Commons (GDC) Hub, n = 472]. B naive, resting dendritic cells, and CD4+ T cells were predicted to occupy a higher percentage in OMMs than in CMs, while the fractions of cytotoxicity-related CD8+ T cells and M1 macrophages were limited, implying an immune-tolerant state and enrichment of immune-inactive pathways (fig. S4C).
Given the higher recurrence and metastasis rates of OMMs compared to CMs (2, 32), we further investigated the transcriptome differences between OMM organoids derived from “primary” OMMs (OMM_OP) and “secondary or metastatic OMMs” (OMM_OS/OLN) (see patient information in table S3) using gene set enrichment analysis (GSEA). The results revealed significant enrichment of the sonic hedgehog (SHH) pathway and ADP ribosylation factor 6 downstream pathway in OMM_OS/OLN, while the Rho-related BTB domain–containing protein 1 guanosine triphosphatase and synthesis of phosphatidic acid pathways were enriched in OMM_OP (P < 0.5; Fig. 3, G and H). Notably, the SHH pathway has been implicated in the metastasis of several epithelial tumors, and its role in OMM progression will be further investigated in future studies (33, 34).
Drug responses of OMM organoids
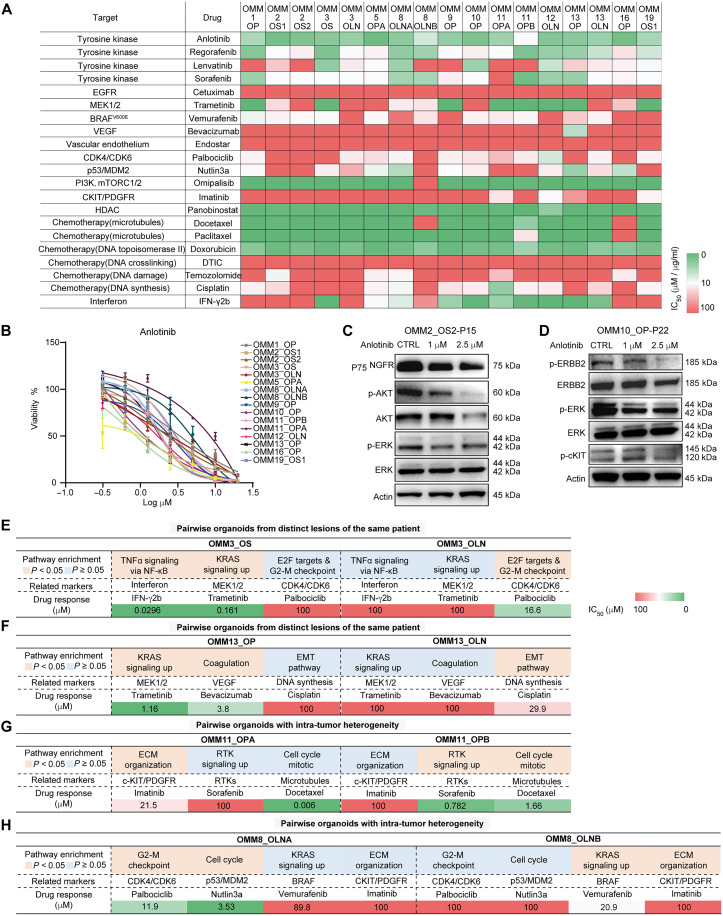
To evaluate the utility of organoids as preclinical models for analyzing drug responses, we conducted an in vitro drug screen to examine the effects of 21 compounds on 17 organoids. The selected drugs encompassed standard-of-care therapies, agents being tested in clinical trials, and those targeting signaling pathways relevant to OMM treatment (Fig. 4A and table S4) (7, 35). We measured cell growth status and determined the half-maximal inhibitory concentration (IC50) values to assess drug responses across different OMM organoids (Fig. 4A and fig. S5A). The biological replications of IC50 values for the 17 screenings exhibited a high correlation (Pearson correlation, r = 0.75, P = 0.0005; fig. S5B), indicating the consistency and reliability of our screening procedures and results.
Fig. 4. Drug responses of OMM organoid lines and their relation to genetics and deregulated expression profile in groups of organoids from the same individual.
(A) Heatmap displaying IC50 values for the tested 21 drugs in OMM organoids, calculated using online GR Calculator. Putative drug targets of the tested compounds are listed on the left. Passage numbers of organoids are provided in table S4. (B) Dose-response curves for OMM organoids treated with the RTK inhibitor anlotinib (n = 4, data presented as the means ± SD). (C) Western blotting results show the dose-dependent inhibition of nerve growth factor receptor (NGFR), phospho-AKT/extracellular signal–regulated kinase (ERK) in OMM2_OS2 organoid treated with 1 and 2.5 μM anlotinib. (D) Western blotting results show the dose-dependent inhibition of phospho-ERBB2/ERK/c-KIT in OMM10 organoid treated with 1 and 2.5 μM anlotinib. (E to H) Tables present both the enriched pathways and related drug responses in pairwise organoid lines. The enrichment pathways were acquired through GO analysis of DEGs in each group. The IC50 values of the drug responses are indicated. OMM3 group (E), OMM13 group (F), OMM11 group (G), and OMM8 group (H).
The drug screening results revealed both notable similarities and differences in the responses to the tested agents among OMM organoids. Particularly noteworthy was the relative sensitivity of organoids (IC50 < 10 μM) to multitarget inhibitors of receptor tyrosine kinases (RTKs), including anlotinib (now tested in a phase 2 clinical trial for OMM treatment; 16 of 17), sorafenib [U.S. Food and Drug Administration (FDA)–approved for hepatocellular carcinoma (HCC); 14 of 17], and regorafenib (approved for HCC; 13 of 17). In addition, two taxanes commonly used in chemotherapy, docetaxel (15 of 17) and paclitaxel (clinically used; 15 of 17), exhibited efficacy against the organoids (Fig. 4, A and B, and fig. S5, C and D). OMMs displayed various RTK alterations, such as CNV amplification of EGFR, ERBB2, FGFR, and METs, as well as activation of RTK pathway or the MAPK pathway regulated by it (Figs. 2, D and E, and 3F). Because of the distinct genomic alterations in different OMMs, the inhibitory effects of multitargeting RTK inhibitors on RTK levels and downstream signaling might be organoid specific. For instance, Western blotting assays demonstrated inhibition of RTK activation in OMM organoids treated with anlotinib, as evidenced by down-regulation of nerve growth factor receptor (NGFR) and decreased phosphorylation levels of extracellular signal–regulated kinase (ERK) and AKT in OMM2_OS2, and reduced phospho–epidermal growth factor receptor 2 and ERK levels in OMM10_OP (Fig. 4, C and D).
In addition, drug responses showed partial correlations between the genome alteration profiles of OMM organoids and their mechanisms of action. For instance, we observed a significant correlation between the IC50 value of palbociclib, an FDA-approved CDK4/CDK6 inhibitor tested in MM xenograft models (4), and CDK4 amplification in OMM organoids (10 of 18 amplification; Pearson correlation, r = 0.61, P < 0.01; fig. S5E). Treatment with palbociclib resulted in the down-regulation of phospho–retinoblastoma protein, a downstream signaling molecule of CDK4 (fig. S5F). Furthermore, we observed a strong correlation between the drug responses to cisplatin and carboplatin, two drugs with similar mechanisms of action (Pearson correlation, r > 0.9, P < 0.001 for OMM11_OPA and OMM19_OS1; r = 0.71 for OMM13_OP; fig. S5G), which has been previously reported in SCCs (36). These findings indicated that organoid lines could serve as reliable in vitro models to provide insights into genetic and drug response heterogeneity.
Among organoid groups derived from the same patients, we observed clear associations between expression profiles and drug responses in chronologically (OMM3/OMM13) and intratumorally (OMM8/OMM11) distinct lesions (Fig. 4, E to H). For example, compared to OMM3_OLN, GO results showed enrichment of TNFα and KRAS signaling in OMM3_OS, which was associated with its sensitivity to interferon-γ (IFN-γ; an immune-related protein) and trametinib (a MAPK kinase inhibitor) (Fig. 4E). Conversely, OMM3_OLN displayed enrichment in E2F targets and G2-M checkpoint and exhibited higher sensitivity to palbociclib than OMM3_OS (Fig. 4E). In the case of OMM11, OMM11_OPB, characterized by CCND1 and VEGFA amplification and enrichment in RTK signaling, showed greater sensitivity to sorafenib than OMM11_OPA. On the other hand, OMM11_OPA, enriched in extracellular matrix (ECM) organization, exhibited a higher response to imatinib [an inhibitor of c-KIT and platelet-derived growth factor receptors (PDGFRs)] than OMM11_OPB (Fig. 4G). The heterogeneous expression and drug response profiles observed during rapid tumor evolution and in different tumor subgroups suggested poorer clinical responses and the potential to improve outcomes through combination therapy.
Our in vitro drug screening using OMM organoids provided evidence that they could serve as preclinical models for analyzing drug responses and their associations with genomic and transcriptional alterations. We observed diverse morphological changes in OMM organoids after treatment with different drugs at concentrations around the IC50. For instance, OMM19_OS1 displayed invasive-like structures following trametinib and panobinostat treatment, while epithelial-like morphology upon treatment with anlotinib and regorafenib (fig. S5H). These morphological changes may be related to the reported variable phenotypes or cellular states of melanoma cells in response to drugs (37, 38).
Drug response of OMM organoid–derived xenografts
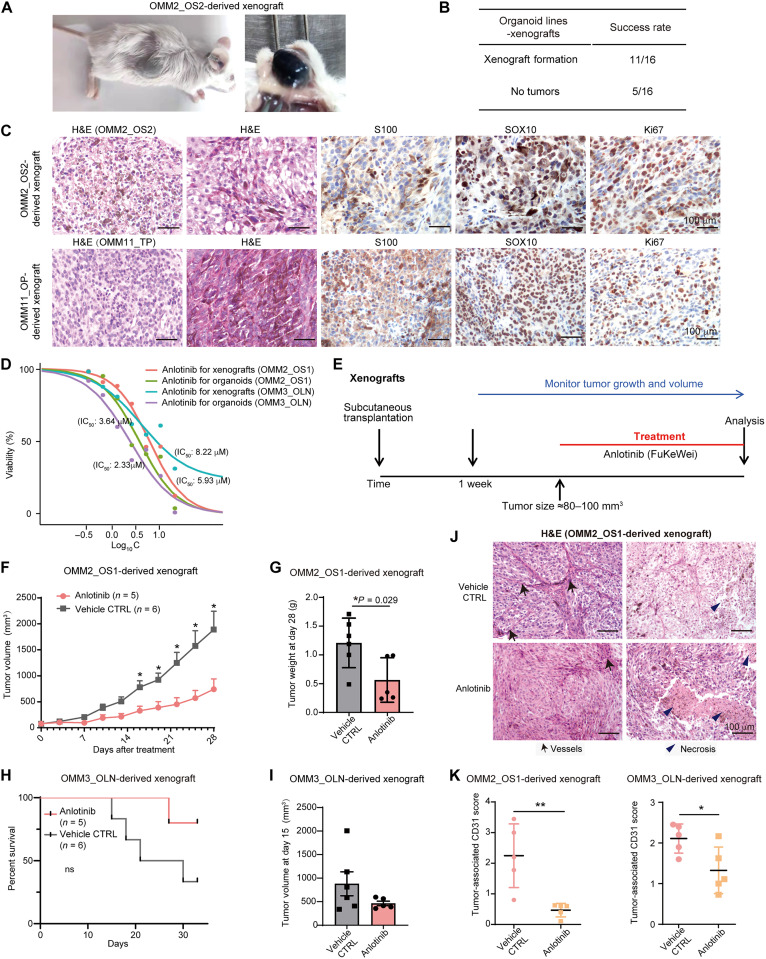
Next, we evaluated the tumorigenic potential of cultured OMM organoids by subcutaneously injecting them in immunodeficient mice to form xenografts. Macroscopically visible tumors were observed after 8 to 12 weeks of transplantation growth (Fig. 5A), with a success rate of 68.8% (11 of 16) for the transplanted organoids (Fig. 5B and table S3). H&E, SOX10, S100, and Ki67 staining of the xenografted tumors revealed their similarity to the original parental OMMs (Fig. 5C).
Fig. 5. Validation of anlotinib response in OMM organoid–derived subcutaneous xenografts in vivo.
(A) Representative images demonstrating the formation of xenografted tumors derived from OMM organoid lines. (B) A summary table of the xenografted tumors for organoid lines (details in table S3). (C) H&E and IHC staining of SOX10, S100, and Ki67 markers in xenografted tumors derived from OMM organoid lines. (D) Dose-response curves for anlotinib treatment in OMM2_OS1-P13 and OMM3_OLN-P17 organoids, using two forms of anlotinib: one for in vitro drug screening (anlotinib, research use only) and the other for in vivo xenografts (FuKeWei, anlotinib product). (E) Overview of the treatment schedule for OMM organoid–derived xenografts. (F) In vivo activity of anlotinib (2 mg/kg) in OMM2_OS1 organoid-derived xenografts, showing the tumor volumes at different time points. Mice were treated with the drug or vehicle daily for 28 days. *P < 0.05, Mann-Whitney test, two-tailed. (G) Bar plots illustrating the tumor weights of xenografts in the treated and control groups. (H) Kaplan-Meier curves for anlotinib- or vehicle-treated mice bearing OMM3_OLN xenografts. ns, not significant. (I) Bar plots presenting the tumor weights of OMM3_OLN xenografts in the anlotinib or vehicle control group. (J) H&E staining results displaying tumor-associated vessels and necrotic areas in OMM organoid–derived xenografts in the anlotinib/vehicle groups. CTRL, control. (K) Quantification of tumor-associated CD31-positive vessels to assess microvascular changes in OMM organoid–derived xenografts in response to anlotinib. OMM2_OS1 (left) and OMM3_OLN (right) (n = 5 animals per group, n = 10 high-power-field tumor areas per animal for statistical scoring). For (F) to (I), n = 5 in the anlotinib group; n = 6 in the vehicle group. Data in (F), (G), (I), and (K) are presented as the means ± SEM, and a two-tailed unpaired Student’s t test was used for the P values. *P < 0.05 and **P < 0.01.
On the basis of our in vitro drug screening data, OMM organoids showed relative sensitivity to anlotinib. Take OMM2_OS1 and OMM3_OLN as examples (Fig. 5D), we validated their responses to FuKeWei (anlotinib, drug product) in subcutaneous xenografts in vivo by administering anlotinib or the vehicle intragastrically when the tumors reached approximately 80 to 100 mm3 (Fig. 5E). In OMM2_OS1 xenografts, we observed a significant reduction in tumor growth 16 days after the first anlotinib treatment, which persisted throughout the experiment (Fig. 5, F and G). Treatment with anlotinib also prolonged the overall survival of OMM3_OLN xenografts (Fig. 5, H and I). Notably, for OMM2_OS1 and OMM3_OLN xenografts, tumor-associated CD31 scores were significantly down-regulated, accompanied by a comparably larger necrotic area, indicating a reduction in microvasculature and live cells in response to anlotinib (Fig. 5, J and K). Collectively, our results demonstrated that the therapeutic effect of anlotinib on OMM organoids in vitro could be recapitulated in an in vivo context.
A coculture system for OMM organoids with autologous immune cells
To facilitate the in vitro investigation of OMM responses to anti–PD-1/PD-L1 therapy, as well as the exploration of resistance mechanisms and combination therapies involving anti–PD-1/PD-L1 antibodies, we developed a coculture system for OMM organoids with autologous PBMCs or TILs (Fig. 6A), based on previously reported methods for immune cell acquisition and coculture (21). The coculture system enabled the assessment of essential indicators for immune cell–mediated cytotoxicity, including the detection of cytokine IFN-γ, TNFα, and cytolytic granzyme B (GZMB) in immune cells, as well as live/dead cell analysis in tumor cells (22).
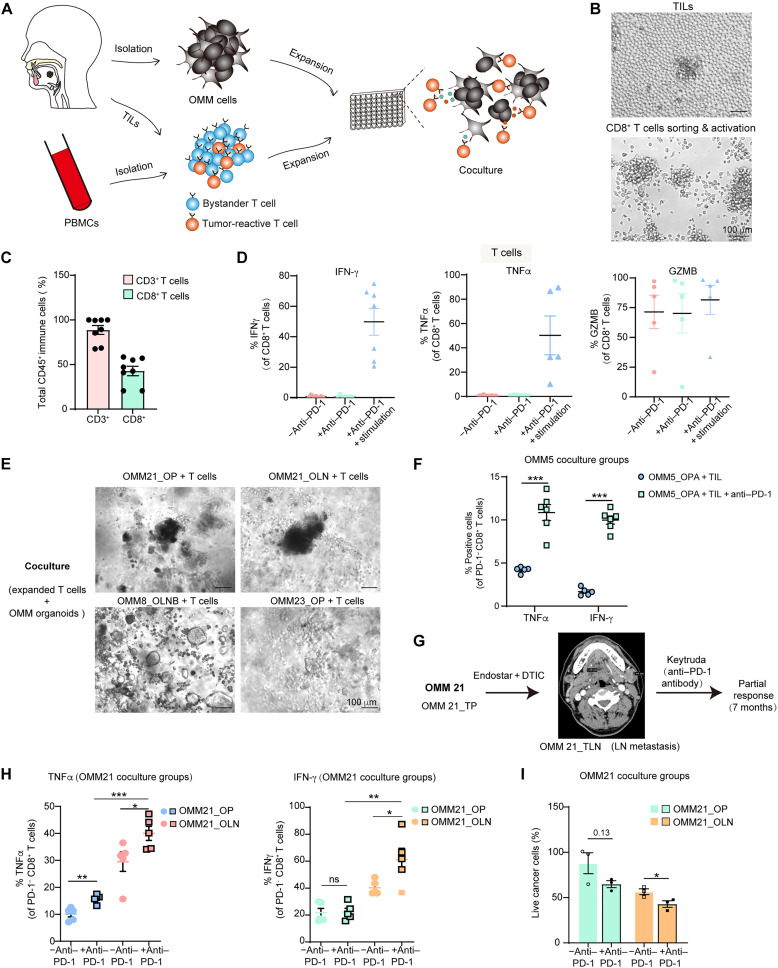
Fig. 6. A coculture system for human OMM organoids and autologous immune cells.
(A) Schematic diagram depicting the generation of organoids, acquisition of autologous immune cells, and establishment of their coculture system. (B) Images showing the morphology of T cells during culture and after activation with anti-CD3 and anti-CD28 antibodies. (C) Percentage of CD3- and CD8-positive cells after 2 weeks of T cell proliferation (n = 8, independent experiments). (D) Percentages of CD8+ T cells positive for granzyme B (GZMB), IFN-γ, and TNFα in the proliferated T cell populations. “−anti–PD-1” refers to no anti–PD-1 antibody (nivolumab) in the culture system, while “+anti–PD-1” indicates the addition of nivolumab in the culture system. “+anti–PD-1 + stimulation” represents the addition of nivolumab and stimulation with PMA and ionomycin in the culture system (n = 7, independent experiments for IFN-γ; n = 5 for TNFα and GZMB). (E) Optical images of small OMM clumps cocultured with T cells. (F) Percentages of PD-1− and CD8+ T cells positive for TNFα and IFN-γ in the OMM5_OPA-P15 organoid and T cell coculture group. (G) Radiation examination and therapeutic schedule for the parental tumors of OMM21 in the clinic. (H) Percentages of PD-1− CD8+ T cells positive for TNFα (left) and IFN-γ (right) in the OMM21_OP-P18 and OMM21_OLN-P11 organoid and autologous T cell coculture group. T cells were acquired after OMM21_TLN TILs cell expansion and CD8+ T microbeads-based cell sorting. (I) Proportions of live cancer cells in OMM21_OP and OLN after coculture with autologous T cells. For (C), (D), (F), (H), and (I), data are presented as the means ± SEM, (F, H, and I) with biologically independent replicates (n = 4 to 6). For (F), (H) and (I), cells were stimulated with PMA and ionomycin. A two-tailed unpaired Student’s t test was used for the P values. *P < 0.05, **P < 0.01, and ***P < 0.001.
Before coculture, immune cells, including PBMCs and TILs, were obtained, expanded, and activated according to established protocols for T cell expansion (see Materials and Methods; Fig. 6, A and B) (21, 39). Although we were only able to acquire 3 to 5 ml of peripheral blood samples from some patients and could not successfully expand TILs from some tumor tissues, we expanded TILs from the corresponding tumor fragments of OMM21_TLN/OMM5_TP/OMM8_TLN. These TILs reached more than 90% CD3+ T cells, with CD8+ T cells comprising over 40% after 2 to 3 weeks of culture (Fig. 6C). Furthermore, we detected CD8+ T cells activities in these groups to ensure functional immune cells before coculture. Stimulation with phorbol myristate acetate (PMA) and ionomycin notably enhanced CD8+ T cell functions in vitro, leading to a substantially higher percentage of effective cytokines expression, such as IFN-γ and TNFα, in CD8+ T cells (Fig. 6D). We applied similar stimulation to T cells for the subsequent coculture system.
We then assessed immune cell–mediated cytotoxicity in the coculture system (Fig. 6E and fig. S6A). CD8+ T cells isolated from TILs and whole PBMCs were activated 48 hours before coculture (Fig. 6B). In the case of OMM5_OPA, the parental tumor had exhibited a clinically favorable response to toripalimab (an anti–PD-1 antibody) with a partial response (PR) lasting over 1 year (table S3). Flow cytometry–based analysis of immune features confirmed a significant increase in the percentages of TNFα and IFN-γ upon nivolumab (an anti–PD-1 antibody) treatment after in vitro coculture of OMM5_OPA with the corresponding T cells, indicating a similar enhanced T cell activities following anti–PD-1 treatment in the coculture system (Fig. 6F).
For OMM21, the patient was treated with endostar (targeting vessels) and dacarbazine (chemo agents) as first-line therapy during the primary stage and with Keytruda (an anti–PD-1 antibody) exhibiting a PR for 7 months at the metastatic stage (Fig. 6G). We successfully expanded TILs from the lymphatic metastasis (OMM21_TLN) and cocultured sorted CD8+ T cells with both OMM21_OP and OMM21_OLN to investigate the differential therapeutic effects of anti–PD-1 on organoids derived from chronologically distinct lesions of OMM21. After nivolumab treatment, the coculture groups showed a significant increase in effective cytokines for T cells, including a notable increase in IFN-γ and TNFα percentages for the OMM21_OLN group and a significant increase in TNFα percentage for the OMM21_OP group (Fig. 6H). These two coculture groups also exhibited reduced live tumor cells, with a significant decrease observed in OMM21_OLN after nivolumab treatment, implying increased cytotoxic activity of immune cells with anti–PD-1 treatment (Fig. 6I). The levels of IFN-γ and TNFα were significantly higher in the OMM21_OLN group compared to that in the OMM21_OP group (P < 0.01; Fig. 6H), suggesting higher T cell activities when cocultured with organoids derived from the parental tumor compared to organoids from chronologically distinct lesions of the same patient. These differences could potentially be attributed to heterogeneous tumor mutation signatures, antigen expressions, the response to IFN-γ signaling, as well as distinct PD-L1 and major histocompatibility complex class I expression levels (40, 41).
In summary, we have developed a coculture and evaluation system for OMM organoids and immune cells, which held promise for analyzing the therapeutic effects of anti–PD-1 therapy or combination therapies. However, CD8+ T cells in PBMCs and TILs exhibited varied tumor recognition and cytotoxic activities, potentially influencing immunotherapeutic responses in the coculture system (21). Although we observed increased effective cytokine expression of T cells in the coculture system following anti–PD-1 treatment, the detected T cell activities were not consistently higher compared to when cultured alone (TNFα, 2 of 5; and IFN-γ, 1 of 5). This discrepancy could be attributed to the diverse antitumor-killing effects of immune cells and the existence of inhibitory elements in the tumor tissues, such as deregulated MAPK signaling, tumor neoantigens, or enrichment of myeloid cells (fig. S6, B and C) (42, 43).
An enrichment of RTK signaling in anti–PD-1–resistant OMMs, with an up-regulation of NGFR in OMM organoids
Previous studies have shown that anti–PD-1/PD-L1 therapy has limited efficacy in MMs. Therefore, we used transcriptome data and established organoids to explore essential factors contributing to OMMs resistance to anti–PD-1 treatment (Fig. 7A). For this analysis, we divided the collected OMMs (10 sequenced, see Materials and Methods) and their corresponding organoids (20 sequenced, see Materials and Methods) into two groups based on the patients’ clinical responses to anti–PD-1 therapy (table S3), namely, an “anti–PD-1–sensitive group” and an “anti–PD-1–resistant group.” Cibersort analysis and multiple immunofluorescent staining (CD8/CD4/CD20/PD-L1/S100B) implied a lower abundance of CD8+ T cells in the anti–PD-1–resistant group compared to the sensitive group (fig. S7, A and B). Consistently, quantitation of immunohistochemistry (IHC) staining in OMM tissues indicated low infiltration of immune cells, including CD8+ and CD4+ T cells (<3%), as well as low PD-L1 expression (<1%) in anti–PD-1–resistant OMM tissues, laterally reflecting their low response to anti–PD-1/PD-L1 therapy (fig. S7C). Subsequently, we analyzed differentially expressed genes (DEGs) to identify associated biological processes in each group. The GO analysis of up-regulated genes in the anti–PD-1–sensitive group revealed an enrichment of T cell receptor (TCR) signaling and CD8 TCR pathway, while the resistant group exhibited an enrichment of ECM organization, interleukin-10 (IL-10) and RTK signaling, and Fos-related antigens pathway (Fig. 7, B and C).
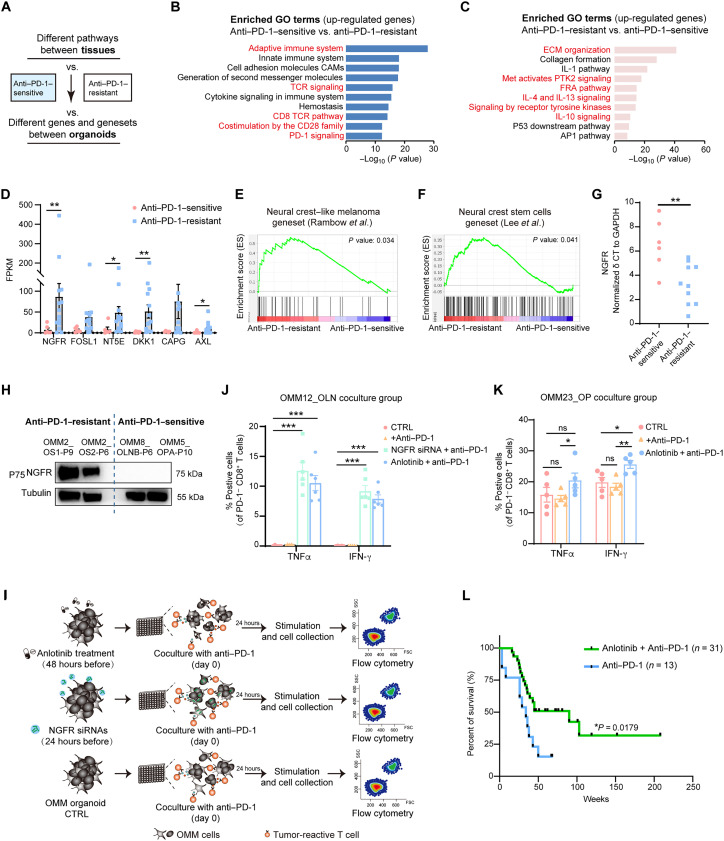
Fig. 7. Combination of anti–PD-1 with inhibition of RTKs or NGFR molecule promotes OMM response to anti–PD-1 therapy.
(A) Flowchart demonstrating the strategies to analyze the transcriptomic differences between “anti–PD-1–sensitive” and “anti–PD-1–resistant” groups. (B and C) Gene pathway analysis of up-regulated DEGs in the anti–PD-1–sensitive (B) and anti–PD-1–resistant (C) groups upon mutual comparison. (D) Average FPKM values for the most variable DEGs in anti–PD-1–resistant OMM organoids compared with anti–PD-1–sensitive OMM organoids. Each dot represents an organoid line. (E and F) GSEA showing enrichment of reported gene sets related to neural crest–like melanoma (E) and neural crest stem cells (NCSCs) (F) in the anti–PD-1–resistant group (for gene set information, see table S5). (G) Delta cycle threshold (δCT) levels (NGFR_ct – GAPDH_ct) for each organoid in the anti–PD-1–resistant and anti–PD-1–sensitive groups. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (H) Western blotting results demonstrating high expression of NGFR in anti–PD-1–resistant organoids. (I) Schematic illustrating the immune combination treatment in the OMM organoids coculture system for exploring therapeutic effects in vitro. Organoids were treated with NGFR siRNAs or anlotinib before coculture with autologous immune cells and anti–PD-1 treatment (nivolumab). Effective activities of immune cells were assessed after the immune combination treatment. SSC, side scatter; FSC, forward scatter. (J) Percentages of PD-1− and CD8+ T cells positive for IFN-γ and TNFα in the NGFR siRNA- or anlotinib-treated OMM12_OLN-P20 organoid and T cell coculture group. (K) Percentages of PD-1− and CD8+ T cells positive for TNFα and IFN-γ in the anlotinib-treated OMM23_OP-P9 organoid and T cell coculture group. For (J) and (K), “+anti–PD-1” denotes the addition of nivolumab in the culture system. A two-tailed unpaired Student’s t test was used for the P values. *P < 0.05, **P < 0.01, and ***P < 0.001. (L) Kaplan-Meier curves for overall survival of patients with OMM with anti–PD-1 antibody treatment alone or a combination treatment comprising anti–PD-1 antibody and anlotinib. Statistics were calculated using the log-rank test.
Considering the beneficial therapeutic response of OMMs to anlotinib, an RTK inhibitor, we focused our investigation on RTK pathways to explore the underlying molecules involved in resistance to anti–PD-1 therapy. Among the DEGs in the organoids of the anti–PD-1–resistant and anti–PD-1–sensitive groups, we observed several highly expressed genes in the anti–PD-1–resistant group (see Materials and Methods for selection criterion), including NGFR and AXL, two members of RTK family (Fig. 7D). Notably, NGFR and AXL have been reported in CMs as potential markers for melanoma cell states known as the “neural crest stem cell (NCSC)–like state” and “undifferentiated state” (40, 44). Melanoma cell states have been associated with metastatic potential and therapeutic sensitivity to MAPK inhibitors and immunotherapies (37, 45). These two states presented a high tolerance to both therapies (38, 44, 46). Consequently, to investigate the phenotypes of OMM organoids in the anti–PD-1–resistant and anti–PD-1–sensitive groups, we performed GSEA using unique transcriptome profiles that define these cell states in CMs (see Materials and Methods and table S5). The GSEA revealed an enrichment of NCSC-related gene sets in the OMM organoids of the anti–PD-1–resistant group, whereas no enrichment was observed in the anti–PD-1–sensitive group (Fig. 7, E and F). Furthermore, we confirmed higher gene and protein expression levels of NGFR in anti–PD-1–resistant OMM organoids using quantitative polymerase chain reaction (qPCR) and Western blotting assays (Fig. 7, G and H).
An improvement on the effective activities of CD8+ T cells for anti–PD-1 therapy by inhibition of RTK signaling and NGFR knockdown in OMMs
Because anti–PD-1–resistant OMMs exhibited enrichment in RTK signaling and the RTK member NGFR, we sought to explore the potential efficacy of combining RTK inhibition with anti–PD-1 therapy. Incidentally, we observed a down-regulation of NGFR protein levels in OMM2 organoids following treatment with anlotinib, an RTK inhibitor (Fig. 4C). To assess the impact of RTK inhibition on anti–PD-1 therapy, we conducted combination therapies using the organoid–immune cell coculture system in vitro. We initiated the inhibition of the RTK family in organoids by either NGFR knockdown or treatment with anlotinib, as previously tested, and subsequently examined whether RTK inhibition could enhance the effectiveness of anti–PD-1 therapy (Fig. 7I).
The corresponding patient of OMM12 was treated with toripalimab after the collection of the OMM12_TLN biopsy with unfavorable response (fig. S7D). Following a 24-hour knockdown of NGFR using small interfering RNAs (siRNAs) or a 48-hour treatment with anlotinib in OMM12_OLN (fig. S7, E and F), we cocultured OMM12_OLN with autologous PBMCs for an additional 24 hours, accompanied by nivolumab treatment (Fig. 7J and fig. S7, G and H). Analysis of immune features using flow cytometry revealed a significant increase in the percentage of IFN-γ+ and TNFα+ cells among CD8+ T cells in the NGFR siRNA and anlotinib treatment group (approximately 10% increase; Fig. 7J). A similar trend, with a significant increase in the percentage of IFN-γ+ and TNFα+ cells among CD8+ T cells, was observed in an anti–PD-1–sensitive organoid, OMM23_OP after being treated with anlotinib and nivolumab (approximately 5% increase; Fig. 7K and fig. S7I). These findings collectively indicated that the combination of anti–PD-1 treatment with RTK family inhibition could enhance the functional activities of T cells. Furthermore, we conducted a retrospective observational study at our hospital and enrolled patients who received either anti–PD-1 treatment alone or a combination of anti–PD-1 therapy with anlotinib. The current statistical analysis demonstrated that patients with OMM receiving a combination therapy exhibited longer overall survival compared to those treated with anti–PD-1 therapy alone (median survival time, 90 weeks versus 34 weeks; P = 0.0179; Fig. 7L).
In summary, our results obtained from the organoid–immune cell coculture system and data from the observational study suggested that combining anti–PD-1 therapy with an RTK inhibitor or NGFR knockdown enhanced the therapeutic efficacy of OMMs. The RTK family, particularly NGFR, potentially emerges as an essential factor in the resistance to anti–PD-1 therapy in OMMs.
DISCUSSION
In this study, we successfully generated 30 organoids that captured the inter- and intratumor heterogeneity of OMMs. These organoids exhibited high concordance in histopathological features and molecular characteristics with parental tumors and human OMMs overall. The lack of approved therapies specific to rare forms of melanoma, such as OMMs, highlights the utility of these organoid models for rapid modeling, drug screening, and preclinical testing of both early and advanced lesions. Through drug screening in OMM organoids, we identified anlotinib, a multitarget RTK inhibitor, as a promising treatment option for OMMs. Moreover, our coculture systems involving OMM organoids and autologous CD8+ T cells or PBMCs revealed that anlotinib treatment or NGFR knockdown in OMM organoids significantly enhanced the immune cell effective activities in the anti–PD-1–resistant group receiving nivolumab treatment.
OMMs exhibit rapid progression and high heterogeneity, prompting us to collect organoid groups from individuals at different disease stages or with intratumor heterogeneity. Genomic analysis of these paired organoids revealed rapid evolution in OMMs, characterized by variations in gene expression profiles, somatic mutations, CNVs, and drug responses. These organoid groups with spatiotemporal tumor heterogeneity have the potential to elucidate mechanisms of tumor evolution, genetic divergence during disease progression, and therapeutic resistance in OMMs through systematic analyses and experimental manipulations of organoid models in future studies.
Immune resistance, both primary and adaptive, contributes considerably to the failure of immunotherapies (47). The limited infiltration of immune cells in “cold tumors” represents an essential obstacle to the effectiveness of anti–PD-1/PD-L1 therapy. Extensive efforts have been made to enhance the response rates of patients with OMM through various immunotherapy combination strategies in clinical trials, including anti–PD-1 + anti–cytotoxic T lymphocyte–associated antigen 4 antibodies, anti–PD-1 antibody + RTK/vascular endothelial growth factor receptor inhibitors, and anti–PD-1 antibody + interferon-α2β (48–50). Early-phase trial data on the combination of anti–PD-1 antibody and RTK inhibitors have shown promising efficacy in OMMs, necessitating further validation in larger prospective randomized studies (51). The improved responsivity may involve alterations in TME, such as influencing the M1/M2 macrophage ratio or T cell sensitivity following RTK inhibition (52, 53). However, because of low levels of infiltrated immune cells in some OMM tissues, our in vitro immunotherapy combination experiments primarily involved coculturing organoids with PBMCs rather than TILs. As the majority of CD8+ T cells in PBMCs are reported to be nontumor specific, our coculture system may not be an ideal model for exploring the mechanisms underlying improved responsivity (22). Future studies can leverage human OMM organoid models and human antigen–specific T cells to develop a high-throughput approach for understanding and predicting mechanisms of action and responses to therapies, leading to discoveries and innovations in next-generation therapy (23).
While a few transplantation models of MMs from patients have been used for preclinical drug testing, the availability of organoid model repository covering heterogeneity and distinct stages of disease progression facilitates the development of molecular subtyping and effective therapies, which would substantially advance our understanding of OMM etiology and disease progression (28, 40, 54). With genetic manipulation, high-throughput drug screening, and integration of TME elements, organoid models offer the potential for further discoveries in this rare subtype of melanoma (14, 15). In vitro genetic engineering of OMM organoids enables the study of rare mutations and a deeper understanding of their oncogenic functions. Long-term low-dose drug treatment in initially sensitive organoid models allows for the exploration of potential mechanisms underlying acquired drug resistance in OMMs (15). In addition, by coculturing organoids with immune or stromal cells in vitro, we could analyze their contributions to therapy resistance in OMMs and identify potential elements involved. This opens up avenues for future OMM research.
In summary, the organoid model repository that we generated for OMMs serves as a valuable resource for studying this rare form of melanoma. These organoids exhibit distinct genomic landscapes and expression profiles compared to CMs. Leveraging these organoids, we were able to gain insights into OMM pathogenesis, develop and predict potential clinical approaches, and lay the foundation for future personalized medicine. This study highlights the power and future potential of organoid-based approaches in OMM research, offering the possibilities for advancing our understanding and treatment of this challenging disease.
MATERIALS AND METHODS
Sample collection
The Institutional Review Boards of Shanghai Jiaotong University School of Medicine and the Ethical Committee of Shanghai Ninth People’s Hospital approved the generation and distribution of organoids, and patient data and tissues were collected with informed consent. Clinical data, including magnetic resonance imaging materials, tumor volume, and treatment regime, were obtained from the medical records system. Patients’ responses to different drugs were evaluated by responsible doctors based on medical records and follow-up therapeutics. The study adhered to all relevant ethical regulations regarding research involving human participants.
Tissue dissociation
Patient material was collected in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 1% penicillin-streptomycin (PS) immediately after surgery and transported to the laboratory on ice. Tissues were washed at least twice with DMEM supplemented with 1% PS and 1× gentamicin-amphotericin solution to remove black secretions. After removing excess muscle or bone, the tissues were cut into small fragments. Biopsies smaller than 3 mm in diameter were used for organoid generation, while larger biopsies were used for genomic analysis and organoid derivation. Random pieces of approximately 5 mm3 were stored at −80°C after treated with the RNAlater Stabilization Solution (Invitrogen) for DNA and RNA isolation. Some pieces were fixed in 4% paraformaldehyde (PFA) for histopathologic analysis, and the remaining fragments were processed for organoid derivation.
Tissue fragments were subjected to mild enzymatic digestion using the Human Tumor Dissociation Kit (Miltenyi Biotec). After 1 min of gentle MACS Dissociator (Miltenyi Biotec) processing, the fragments were placed on an orbital shaker at 37°C for 20 to 30 min. Tumor cells were collected by centrifugation at 1500 rpm for 5 min. Approximately 1 × 104 cells were mixed with Matrigel (Corning) and seeded into each well of prewarmed 24-well flat-bottom cell culture plates (Corning Costar).
OMM organoid culture
OMM organoid generation and culture
OMM organoids were generated and cultured on the basis of published protocols with minor modifications. Organoids were grown in Advanced DMEM (Life Technologies) supplemented with 1× B27 supplement (Life Technologies), 1× N2 supplement (Life Technologies), 1.25 mM N-acetyl-l-cysteine (Sigma-Aldrich), 10 mM nicotinamide (Sigma-Aldrich), human epidermal growth factor (50 ng/ml; PeproTech), human fibroblast growth factor 2 (5 ng/ml; PeproTech), 50 nM endothelin-3 (MedChemExpress), 500 nM A83-01 (Tocris Bioscience), 1 μM forskolin (R&D Systems), 10 μM Y-27632 (MedChemExpress), 0.3 μM CHIR-99021 (Sigma-Aldrich), human stem cell factor (50 ng/ml; PeproTech), Primocin (2.5 mg/ml; InvivoGen), 8% R-spondin–conditional medium (produced using media collected from human embryonic kidney–293 cell lines expressing recombinant R-spondin-1, provided by Lijian Hui laboratory), and 2% fetal bovine serum (FBS) (Life Technologies).
Passage of OMM organoids
OMM cells began to proliferate approximately 1 week after initial plating (passage 0, “P0”) and were ready for passage after 1 to 3 weeks, depending on their growth conditions. For passaging, organoids were collected from the plate by disrupting the Matrigel droplets with a P1000 pipette, collecting and washing them in a 15-ml centrifuge tube. Matrigel was depolymerized using cell recovery solution (Corning), and the released cell pellets were resuspended in 1 ml of DMEM for mechanical digestion. The cell suspension was gently pipetted up and down for around 20 to 50 times using a P200 pipette while closely monitoring the sensitivity of OMM cells to mechanical force, or they would grow much slower or become senescent after relatively harsh treatment. The centrifuged pellets were resuspended with Matrigel and divided into a 24-well suspension plate (50 μl per well). Once the Matrigel droplets polymerized, 1 ml of culture medium was added, and the medium was changed every 4 days.
Freeze and recovery of OMM organoids
To freeze organoids, Matrigel was dissolved with cell recovery solution, and the cell clusters were pelleted. The pellets were resuspended in freezing medium (10% dimethyl sulfoxide + 90% FBS) and slowly frozen at −80°C overnight, followed by storage in liquid nitrogen. The frozen cells were recoverable using a standard liquid nitrogen cell-recovery protocol and could be embedded in Matrigel. Mycoplasma PCR analysis (Mycoplasma PCR Detection Kit, Beyotime) confirmed that all cultured organoids used in the experiments were mycoplasma negative.
Fluorescence in situ hybridization
FISH was performed as previously described (4). CDK4 and TERT amplification tests were conducted on formalin-fixed and paraffin-embedded tissue sections using commercial probes: GSP CDK4/CSP 12 Dual Color Probe and GSP TERT/GSP PDGFRB Dual Color Probe (AccuPa+h). The experimental procedures followed the manufacturer’s instructions.
Organoid xenograft studies
The Institutional Animal Care and Use Committee of the Ninth People’s Hospital of the Shanghai JiaoTong University School of Medicine approved all animal experimental procedures. To establish xenografts, organoids were washed twice with phosphate-buffered saline (PBS) on the day of transplantation. Subsequently, organoids were gently disrupted using a P200 pipette and divided into smaller masses. Then, 2 to 3 million cells suspended in 150 μl of a 50% matrigel/PBS mixture were subcutaneously injected into female NCG mice (NOD/ShiLtJGpt-Prkdcem26Cd52Il2rgem26Cd22/Gpt) (GemPharmatech) aged between 6 and 8 weeks. After 2 to 4 months, mice were euthanized by cervical dissociation, and tumors were excised and fixed overnight in 4% PFA.
For in vivo drug studies, three successfully engrafted organoid-derived xenografts were minced into smaller pieces of approximately 30 mm3 (3 mm by 3 mm by 3 mm) and subcutaneously injected into the flanks of 5-week-old nude mice (Slarc) for subsequent studies. Once the tumor volume reached 80 to 100 mm3, the mice were randomly divided into two groups: (i) control group with PBS administration (100 μl, intragastric administration, once daily) and (ii) anlotinib group (2 mg/kg, intragastric administration, once daily). The health status of the mice and tumor growth were monitored daily. Tumor volume and mouse body weight were recorded twice per week. The mice were euthanized 28 days after drug intervention or when the tumor volume reached 2000 mm3. Tumor tissues were harvested for measurement of tumor volume and fixed for histological examination. Tumor size was measured using a caliper, and tumor volume was calculated using the formula: tumor volume = 0.5 × length × (width)2, as previously described.
Drug assays for OMM organoids
OMM organoids were harvested and dissociated into single cells, filtered using a 40-μm nylon cell strainer, and then centrifuged and resuspended in 1 ml of advanced DMEM/F12 medium. The cells were seeded in standard 96-well cell culture plates (Corning) at a density of 1 × 104 cells per well. After solidification of the Matrigel, 110 μl of human OMM organoid media was overlaid. Three to 4 days after seeding, 50 μl of medium was removed and replaced with 55 μl of drug-containing human OMM organoid medium.
In brief, we selected 21 compounds and vehicle controls for drug screening (complete drug information in table S4). Each screen was conducted in four replicates, starting with appropriate concentrations determined in pre-experiments. The drugs were added at the designated time, and, after 96 hours, adenosine 5′-triphosphate levels were measured using the CellTiter-Glo 3D Reagent (Promega) according to the manufacturer’s instructions. Luminescence was measured using a Spark multimode microplate reader (Tecan). The results were normalized to the vehicle control (100%). Kill curves were generated using GraphPad software, and lines were fitted using the “log (inhibitor) versus 4 response variable slopes” option. The IC50 value was calculated using the online GR Calculator and assigned value of 100 when over 100.
Coculture and characterization methods
PBMC culture
The PBMC fraction was isolated from peripheral blood using the Lympholyte-H Cell Separation Media (Cedarlane) and cryopreserved for later use. The culture medium for PBMCs consisted of RPMI 1640 (Corning), supplemented with 2 mM ultraglutamine I, 1% PS solution, 25 mM Hepes (pH 7.2), 5.5 × 10−5 M β-mercaptoethanol, IL-2 (6000 IU/ml; Sigma-Aldrich and SL Pharm), and 10% inactivated serum (Gibco) (referred to as “T cell expansion medium”). CD3 monoclonal antibody (30 ng/ml) and CD28 antibody (clone CD28.2, eBioscience) were added for T cell activation at least 48 hours before coculture (referred to as “T cell activation medium”).
TIL culture
For TIL culture, we followed published protocols (21, 39). Briefly, during OMM tissues processing into organoids, small fragments were saved before and after digestion for TIL culture. The fragments were placed in each well of a 24-well tissue culture plate with 2 ml of T cell expansion medium. The plates were then placed in a humidified 37°C incubator with 5% CO2 for culture, and half of the medium was replaced every 4 days. After 1 to 2 weeks, once lymphocyte growth was evident, the cells were transferred to 25-cm3 flasks for expansion.
Coculture of lymphocytes and OMM organoids
To evaluate the blocking effect of the anti–PD-1 antibody on T cells, we conducted coculture assays of OMM organoids with CD8+ TILs and CD45+ PBMCs. CD8+ TILs and CD45+ PBMCs were sorted with CD8 and CD45 MicroBeads (Miltenyi Biotec), respectively. On the day of coculture, we digested the organoids and resuspended the small cell clumps or single cells in T cell activation medium. CD8+ TILs or CD45+ PBMCs were seeded at a density of 105 cells per well and stimulated with the dissociated organoids at an effector-to-target ratio of 5:1 or 10:1. Cocultures were carried out for 24 hours in the presence of T cell activation medium, using 96-well round-bottom ultralow attachment microplates (Corning). In the experimental group, nivolumab (40 to 80 ng/μl; anti–PD-1 antibody) was added. For organoids subjected to siRNA treatment for 24 hours and anlotinib treatment for 48 hours, the entire coculture system for all groups was extended to around 36 hours.
Flow cytometry
To determine the proportions of CD8+ or CD45+ lymphocytes, lymphatic cells were washed with PBS and stained with FVS440 (BD Biosciences) for live/dead cell discrimination. After washing with Stain buffer (BD Biosciences) and blocking with Human Fc Block (BD Biosciences), the cells were stained with anti–CD45-Peridinin chlorophyll protein-Cyanine5.5 (PerCP-Cy5.5), anti–CD3-Alexa700, anti–CD4–allophycocyanin (APC)–cy7, and anti–CD8–fluorescein isothiocyanate (FITC) antibodies (BD Biosciences) for 30 min at 4°C.
To evaluate tumor reactivity, Golgi-Plug containing the leukocyte activation cocktail (1:500, BD Biosciences, containing PMA and ionomycin) was added 8 to 10 hours before cell harvest. The cells were first stained with FVS440, Fc-blocked, and then stained with membrane anti–CD8-FITC and anti–PD-1–phycoerythrin–cy7 antibodies. The cell membranes were then permeabilized using the Cytofix/Cytoperm Fixation/Permeabilization Kit (BD Biosciences) according to the manufacturer’s instructions. Intracellular antibodies, including anti–IFN-γ–APC, anti–GZMB-AF700, or anti–TNFa-R719 (BD Biosciences), were added for 30 min at 4°C. To evaluate the viability of the OMM cells, the OMM cells were stained with carboxyfluorescein diacetate succinimidyl ester–FITC (BD Biosciences) before coculture and then stained with anti–CD45-PerCP-Cy5.5 antibodies and FVS440 for the culture system to analyze the percentage of live tumor cells.
Sample acquisition was performed using the LSR AriaSORP III or LSR Fortessa (BD Biosciences) system, and the data were analyzed using FlowJo v.10.6.0. The antibodies information is listed in table S6.
Histology and immunofluorescent staining
For immunofluorescent staining, the organoids were fixed with 4% PFA for 1 hour, subsequently permeabilized with 0.3% Triton X-100 (Sigma-Aldrich) for 15 min, followed by three washes with PBS, and blocked with 3% bovine serum albumin (BSA) (Solarbio) for 1 hour. Primary antibodies (refer to table S6) were incubated with the organoids overnight at 4°C. Afterward, fluorescence-conjugated secondary antibodies were applied to the organoids for 1 hour at room temperature in the dark. Last, the nuclei were stained with 4′,6-diamidino-2-phenylindole for 5 min. Zeiss 800 confocal microscopes were used to capture the images.
For H&E and IHC staining, tissues and organoids were fixed overnight at room temperature in 4% PFA. The fixed samples were then processed using standard protocols, including dehydration, clearing, and embedding in paraffin. To optimize visualization in paraffin blocks, additional drops of eosin were added to the organoids during the 100% ethanol step. Images were observed using a Zeiss Z5 microscope.
Western blotting
Western blotting was performed following a routine protocol. OMM organoids, treated with drugs for approximately 48 hours, were lysed in radioimmunoprecipitation assay lysis buffer. An equal amount of total protein lysates was separated by 4 to 20% SDS–polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane (Millipore). The membrane was then blocked with 5% (w/v) BSA (Solarbio) and incubated overnight with primary antibodies at 4°C, followed by incubation with secondary antibodies for 1 hour. Protein bands were visualized using Western Bright ECL Chemiluminescent HRP Substrate (advansta). All antibodies are listed in table S6.
Reverse transcription qPCR and siRNA transfection
Total RNA was isolated from cells using RNAiso Plus (TaKaRa). Reverse transcription into cDNA was performed using the PrimeScript RT reagent Kit (TaKaRa) with 0.8 μg of RNA. qPCR was carried out using SYBR Premix Ex Taq (TaKaRa) on a Roche real-time PCR system. The primer sequences are listed in table S7. For siRNA transfection in a 24-well plate, 100 nM siRNA was transfected with 2 μl of Lipofectamine 3000 (Invitrogen) in Opti-MEM (Gibco) for each well. RNA and protein expression levels were detected after transfection for 30 and 72 hours, respectively.
Transmission electron microscopy
Organoids in culture medium were washed twice with PBS and fixed with 2.5% glutaraldehyde overnight. Subsequently, they were washed three times with 0.1 M phosphate buffer and postfixed with 2% osmium tetroxide. Dehydration was performed using a step-up grade series of ethanol, followed by clearing in acetone and infiltration in fresh 100% resin. The samples were then polymerized in an oven at 60°C for 48 hours. These procedures were conducted in 1.5-ml tubes after the organoids sedimented at the bottom. Ultrathin sections (70 nm) were prepared, stained with uranyl acetate and lead citrate, and examined by electron microscopy (FEI Tecnai G2 Spirit).
RNA sequencing
RNA-seq was conducted on a Illumina NovaSeq 6000 platform (SEQUANTA) in PE150 mode. Quality control was performed using fastp. The reads were aligned to the human reference genome (GRCh38) using STAR (55, 56). FeatureCounts was used to count the reads mapping to genomic features (GENCODE v38) (57). Transcripts per million and FPKM (fragments per kilobase of exon per million fragments mapped) values were calculated using StringTie (58). Only protein-coding genes were included for further study, and genes with read counts less than 10 in over half of the samples were filtered out.
PCA was performed using R’s prcomp(). The scatterplot of sample projection onto the first two principal components was plotted.
GO analysis was conducted for DEGs (fold change of FPKM and FPKM of at least one sample > 5) in pairwise samples using the GSEA website (Broad Institute, http://software.broadinstitute.org/gsea/msigdb/annotate.jsp). Significance was indicated by a false discovery rate q value of <0.05.
GSEA analysis was performed using the GSEA software (59) (www.broadinstitute.org/gsea/) by analyzing the FPKM values of two groups in comparison.
The anti–PD-1–resistant group and anti–PD-1–sensitive group were determined on the basis of the patients’ response to anti–PD-1 therapies. Patients who showed PR to anti–PD-1 treatment for 1 year were considered sensitive, while patients with stable or progressive diseases were considered resistant. Additional information on patients is provided in table S3.
The up-regulated genes in DEGs for organoids of the anti–PD-1–resistant and anti–PD-1–sensitive groups were selected on the basis of the following criterion: average FPKM of 14 resistant organoids from highest to lowest, FPKM > 10 (>10 resistant samples), and FPKM < 10 (≥4 of the 6 sensitive samples). The listed genes in figures were either RTK- or melanoma-associated.
RNA-seq data of acral and CM were downloaded from Weiss et al. (31). The provided counts value was used for comparison in our analyses.
For online Cibersort analysis, RNA-seq data of TCGA_SKCM (472 samples) were downloaded from GDC Hub (https://gdc.xenahubs.net) for analysis (60). Normal samples ending with “11A” were excluded, and resting samples were classified into two clusters. Primary samples with a name of “01A” and secondary samples with “06A” were involved.
Whole-exon sequencing
Libraries of OMM organoids, parental tumor tissues, and blood samples were prepared using the SureSelect Human All Exon V6 Capture Kit (Agilent Technologies). WES was performed on an Illumina NovaSeq 6000 system for 150–base pair paired-end sequencing (Mingma Technologies Co. Ltd., Shanghai, China). The average sequencing depth was 200× for organoids and tumor tissues and 50× for blood samples or para-tumor normal samples. The sequence data were aligned to the human genome (GRCh38) using Burrows-Wheeler Aligner (61) and transformed to a coordinate-sorted binary file Binary Alignment/Map format (BAM). MuTect2 (62) was used to process BAM files and detect point mutations, and Variant Effect Predictor version 104 was used to annotate gene transcripts in GENCODE v38. Mutation files were converted to MAF format using vcf2maf (version 1.6.21) and visualized using the R package maftools.
SNV calling
High-confidence somatic mutations were obtained by filtering out germline mutations and applying multiple thresholds, including filtration of variants in dbSNP138 and 1000 genomes databases, low base quality (<20), low mapping quality (<30), and low allele frequency of alteration (<0.05).
CNV detection
Somatic CNV detection was performed using Control-FREEC (v11.6) with matched tumor and normal BAM files (63). Multiple thresholds, including read counts (>50) and break point (>1.2), were applied. Genomic regions with copy number amplification (≥3) or deletion (≤1) were defined as gain or loss, respectively.
Statistical analyses
Statistical analyses, including two-sided Student’s t test and Fisher’s exact test, were conducted using Microsoft Excel or GraphPad Prism 8 software. The data are presented as the means ± SD or the means ± SEM, and P values were calculated. GSEA was used for pairwise comparisons on the basis of expression differences. Kaplan-Meier curves with log-rank statistics were used for comparing overall survival. For additional statistical significance values and sample sizes in graphs, refer to the figure legends and Materials and Methods for more details.
Supplementary Material
Acknowledgments:
We thank R. Cui and M. Qiu for suggestions on the manuscript and revision; Z. Tian and Q. Lin for pathological examination; H. Dong and Y. Huang for assistance with the WES and RNA-seq analysis; Y. Bai, L. Cui, Y. Wang, and Shanghai Key Laboratory of Translational Medicine on Ear and Nose diseases (no. 14DZ2260300) for assistance with flow cytometry; and Electron Microscopy System at Shanghai Institute of Precision Medicine for assistance in TEM images and other members of the Shuyang Sun laboratory for their valuable suggestions.
Funding: This work was supported by the Key Program of National Natural Science Foundation of China (82030085), the National Natural Science Foundation of China (82103051 and 81872199), the National Key Research and Development Program of China (2017YFC0908500), Innovative Research Team of High-level Local Universities in Shanghai (SHSMU-ZLCX20212300 and SSMU-ZLCX20180500), “Shuguang Program” supported by Shanghai Education Development Foundation and Shanghai Municipal Education Commission (19SG13), China National Postdoctoral Program for Innovative Talents (BX2021189), China Postdoctoral Science Foundation (2021M692113), Shanghai Clinical Research Center for Oral Diseases (19MC1910600), Shanghai Municipal Key Clinical Specialty (shslczdzk01601), Shanghai’s Top Priority Research Center (2022ZZ01017), and CAMS Innovation Fund for Medical Sciences (CIFMS) (2019-I2M-5-037).
Author contributions: Conceptualization: S.S. and L.S. Methodology: S.S. and L.S. Investigation—experiments and bioinformatic analyses: L.S. X.K., and C.W. Investigation—sample and clinical data collection: C.W., X.K., H.J., G.Y., R.W., and L.S. Investigation—pathology diagnosis: H.J., G.Y., and S.S. Writing—original draft: L.S. and S.S. Writing—review and editing: X.K. and S.S. Funding acquisition: S.S. and L.S. Supervision: S.S.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Further information and requests for organoids models, other resources, and reagents should be directed to and will be fulfilled by the lead contact, S.S. (sunshuyang@sjtu.edu.cn). The WES and RNA-seq raw data generated in this study were deposited in the Genome Sequence Archive (GSA) in the BIG Data Center (https://ngdc.cncb.ac.cn/gsa-human/), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession numbers HRA004689 and HRA004709.
Correction (8 November 2023): Due to a production error, the x axis labels in Fig. 7G were duplicated to both read, “anti-PD-1-sensitive,” and the labels at the top of the Western blot in Fig. 7H were duplicated to both read, “anti-PD-1-resistant.” Fig. 7G has been corrected so that the label on the right reads, “anti-PD-1-resistant.” Fig. 7H has been corrected so that the label at the top right of the Western “anti-PD-1-sensitive.” Additionally, the extraneous text, “DTIC, dacarbazine” was incorrectly added to the Fig. 5 caption during copyediting and has been removed.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Legends for tables S1 to S7
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S7
REFERENCES AND NOTES
- 1.Spencer K. R., Mehnert J. M., Mucosal melanoma: Epidemiology, biology and treatment. Cancer Treat. Res. 167, 295–320 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Tyrrell H., Payne M., Combatting mucosal melanoma: Recent advances and future perspectives. Melanoma Manag. 5, MMT11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lian B., Cui C. L., Zhou L., Song X., Zhang X. S., Wu D., Si L., Chi Z. H., Sheng X. N., Mao L. L., Wang X., Tang B. X., Yan X. Q., Kong Y., Dai J., Li S. M., Bai X., Zheng N., Balch C. M., Guo J., The natural history and patterns of metastases from mucosal melanoma: An analysis of 706 prospectively-followed patients. Ann. Oncol. 28, 868–873 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Zhou R., Shi C., Tao W., Li J., Wu J., Han Y., Yang G., Gu Z., Xu S., Wang Y., Wang L., Wang Y., Zhou G., Zhang C., Zhang Z., Sun S., Analysis of mucosal melanoma whole-genome landscapes reveals clinically relevant genomic aberrations. Clin. Cancer Res. 25, 3548–3560 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Hayward N. K., Wilmott J. S., Waddell N., Johansson P. A., Field M. A., Nones K., Patch A.-M., Kakavand H., Alexandrov L. B., Burke H., Jakrot V., Kazakoff S., Holmes O., Leonard C., Sabarinathan R., Mularoni L., Wood S., Xu Q., Waddell N., Tembe V., Pupo G. M., De Paoli-Iseppi R., Vilain R. E., Shang P., Lau L. M. S., Dagg R. A., Schramm S.-J., Pritchard A., Dutton-Regester K., Newell F., Fitzgerald A., Shang C. A., Grimmond S. M., Pickett H. A., Yang J. Y., Stretch J. R., Behren A., Kefford R. F., Hersey P., Long G. V., Cebon J., Shackleton M., Spillane A. J., Saw R. P. M., López-Bigas N., Pearson J. V., Thompson J. F., Scolyer R. A., Mann G. J., Whole-genome landscapes of major melanoma subtypes. Nature 545, 175–180 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Nassar K. W., Tan A. C., The mutational landscape of mucosal melanoma. Semin. Cancer Biol. 61, 139–148 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seth R., Messersmith H., Kaur V., Kirkwood J. M., Kudchadkar R., Quade J. L. M., Provenzano A., Swami U., Weber J., Alluri K. C., Agarwala S., Ascierto P. A., Atkins M. B., Davis N., Ernstoff M. S., Faries M. B., Gold J. S., Guild S., Gyorki D. E., Khushalani N. I., Meyers M. O., Robert C., Santinami M., Sehdev A., Sondak V. K., Spurrier G., Tsai K. K., van Akkooi A., Funchain P., Systemic therapy for melanoma: ASCO guideline. J. Clin. Oncol. 38, 3947–3970 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Zhang S., Zhang J., Guo J., Si L., Bai X., Evolving treatment approaches to mucosal melanoma. Curr. Oncol. Rep. 24, 1261–1271 (2022). [DOI] [PubMed] [Google Scholar]
- 9.Yu J., Yan J., Guo Q., Chi Z., Tang B., Zheng B., Yu J., Yin T., Cheng Z., Wu X., Yu H., Dai J., Sheng X., Si L., Cui C., Bai X., Mao L., Lian B., Wang X., Yan X., Li S., Zhou L., Flaherty K. T., Guo J., Kong Y., Genetic aberrations in the CDK4 pathway are associated with innate resistance to PD-1 blockade in chinese patients with non-cutaneous melanoma. Clin. Cancer Res. 25, 6511–6523 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Shui I. M., Liu X. Q., Zhao Q., Kim S. T., Sun Y., Yearley J. H., Choudhury T., Webber A. L., Krepler C., Cristescu R., Lee J., Baseline and post-treatment biomarkers of resistance to anti-PD-1 therapy in acral and mucosal melanoma: An observational study. J. Immunother. Cancer 10, e004879 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao L., Qi Z., Zhang L., Guo J., Si L., Immunotherapy in acral and mucosal melanoma: Current status and future directions. Front. Immunol. 12, 680407 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rebecca V. W., Somasundaram R., Herlyn M., Pre-clinical modeling of cutaneous melanoma. Nat. Commun. 11, 2858 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patton E. E., Mueller K. L., Adams D. J., Anandasabapathy N., Aplin A. E., Bertolotto C., Bosenberg M., Ceol C. J., Burd C. E., Chi P., Herlyn M., Holmen S. L., Karreth F. A., Kaufman C. K., Khan S., Kobold S., Leucci E., Levy C., Lombard D. B., Lund A. W., Marie K. L., Marine J.-C., Marais R., Mahon M. M., Robles-Espinoza C. D., Ronai Z. A., Samuels Y., Soengas M. S., Villanueva J., Weeraratna A. T., White R. M., Yeh I., Zhu J., Zon L. I., Hurlbert M. S., Merlino G., Melanoma models for the next generation of therapies. Cancer Cell 39, 610–631 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tuveson D., Clevers H., Cancer modeling meets human organoid technology. Science 364, 952–955 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Veninga V., Voest E. E., Tumor organoids: Opportunities and challenges to guide precision medicine. Cancer Cell 39, 1190–1201 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Yao Y., Xu X., Yang L., Zhu J., Wan J., Shen L., Xia F., Fu G., Deng Y., Pan M., Guo Q., Gao X., Li Y., Rao X., Zhou Y., Liang L., Wang Y., Zhang J., Zhang H., Li G., Zhang L., Peng J., Cai S., Hu C., Gao J., Clevers H., Zhang Z., Hua G., Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell Stem Cell 26, 17–26.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Kawasaki K., Toshimitsu K., Matano M., Fujita M., Fujii M., Togasaki K., Ebisudani T., Shimokawa M., Takano A., Takahashi S., Ohta Y., Nanki K., Igarashi R., Ishimaru K., Ishida H., Sukawa Y., Sugimoto S., Saito Y., Maejima K., Sasagawa S., Lee H., Kim H.-G., Ha K., Hamamoto J., Fukunaga K., Maekawa A., Tanabe M., Ishihara S., Hamamoto Y., Yasuda H., Sekine S., Kudo A., Kitagawa Y., Kanai T., Nakagawa H., Sato T., An organoid biobank of neuroendocrine neoplasms enables genotype-phenotype mapping. Cell 183, 1420–1435.e21 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Lõhmussaar K., Oka R., Valle-Inclan J. E., Smits M. H. H., Wardak H., Korving J., Begthel H., Proost N., van de Ven M., Kranenburg O. W., Jonges T. G. N., Zweemer R. P., Veersema S., van Boxtel R., Clevers H., Patient-derived organoids model cervical tissue dynamics and viral oncogenesis in cervical cancer. Cell Stem Cell 28, 1380–1396.e6 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Shi X., Li Y., Yuan Q., Tang S., Guo S., Zhang Y., He J., Zhang X., Han M., Liu Z., Zhu Y., Gao S., Wang H., Xu X., Zheng K., Jing W., Chen L., Wang Y., Jin G., Gao D., Integrated profiling of human pancreatic cancer organoids reveals chromatin accessibility features associated with drug sensitivity. Nat. Commun. 13, 2169 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dijkstra K. K., Cattaneo C. M., Weeber F., Chalabi M., van de Haar J., Fanchi L. F., Slagter M., van der Velden D. L., Kaing S., Kelderman S., van Rooij N., van Leerdam M. E., Depla A., Smit E. F., Hartemink K. J., de Groot R., Wolkers M. C., Sachs N., Snaebjornsson P., Monkhorst K., Haanen J., Clevers H., Schumacher T. N., Voest E. E., Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell 174, 1586–1598.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cattaneo C. M., Dijkstra K. K., Fanchi L. F., Kelderman S., Kaing S., van Rooij N., van den Brink S., Schumacher T. N., Voest E. E., Tumor organoid–T-cell coculture systems. Nat. Protoc. 15, 15–39 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Z., Van der Jeught K., Fang Y., Yu T., Li Y., Ao Z., Liu S., Zhang L., Yang Y., Eyvani H., Cox M. L., Wang X., He X., Ji G., Schneider B. P., Guo F., Wan J., Zhang X., Lu X., An organoid-based screen for epigenetic inhibitors that stimulate antigen presentation and potentiate T-cell-mediated cytotoxicity. Nat. Biomed. Eng. 5, 1320–1335 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dao V., Yuki K., Lo Y. H., Nakano M., Kuo C. J., Immune organoids: From tumor modeling to precision oncology. Trends Cancer 8, 870–880 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neal J. T., Li X., Zhu J., Giangarra V., Grzeskowiak C. L., Ju J., Liu I. H., Chiou S.-H., Salahudeen A. A., Smith A. R., Deutsch B. C., Liao L., Zemek A. J., Zhao F., Karlsson K., Schultz L. M., Metzner T. J., Nadauld L. D., Tseng Y.-Y., Alkhairy S., Oh C., Keskula P., Mendoza-Villanueva D., De La Vega F. M., Kunz P. L., Liao J. C., Leppert J. T., Sunwoo J. B., Sabatti C., Boehm J. S., Hahn W. C., Zheng G. X. Y., Davis M. M., Kuo C. J., Organoid modeling of the tumor immune microenvironment. Cell 175, 1972–1988.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ascierto P. A., Accorona R., Botti G., Farina D., Fossati P., Gatta G., Gogas H., Lombardi D., Maroldi R., Nicolai P., Ravanelli M., Vanella V., Mucosal melanoma of the head and neck. Crit. Rev. Oncol. Hematol. 112, 136–152 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Driehuis E., Kretzschmar K., Clevers H., Establishment of patient-derived cancer organoids for drug-screening applications. Nat. Protoc. 15, 3380–3409 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Raposo G., Marks M. S., Melanosomes–dark organelles enlighten endosomal membrane transport. Nat. Rev. Mol. Cell Biol. 8, 786–797 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thawani R., Kim M. S., Arastu A., Feng Z., West M. T., Taflin N. F., Thein K. Z., Li R., Geltzeiler M., Lee N., Fuller C. D., Grandis J. R., Floudas C. S., Heinrich M. C., Hanna E., Chandra R. A., The contemporary management of cancers of the sinonasal tract in adults. CA Cancer J. Clin. 73, 72–112 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Jaarsveld R. H., Kops G. J. P. L., Difference makers: Chromosomal instability versus aneuploidy in cancer. Trends Cancer 2, 561–571 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Mort R. L., Jackson I. J., Patton E. E., The melanocyte lineage in development and disease. Development 142, 620–632 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss J. M., Hunter M. V., Cruz N. M., Baggiolini A., Tagore M., Ma Y., Misale S., Marasco M., Simon-Vermot T., Campbell N. R., Newell F., Wilmott J. S., Johansson P. A., Thompson J. F., Long G. V., Pearson J. V., Mann G. J., Scolyer R. A., Waddell N., Montal E. D., Huang T.-H., Jonsson P., Donoghue M. T. A., Harris C. C., Taylor B. S., Xu T., Chaligné R., Shliaha P. V., Hendrickson R., Jungbluth A. A., Lezcano C., Koche R., Studer L., Ariyan C. E., Solit D. B., Wolchok J. D., Merghoub T., Rosen N., Hayward N. K., White R. M., Anatomic position determines oncogenic specificity in melanoma. Nature 604, 354–361 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klemen N. D., Wang M., Rubinstein J. C., Olino K., Clune J., Ariyan S., Cha C., Weiss S. A., Kluger H. M., Sznol M., Survival after checkpoint inhibitors for metastatic acral, mucosal and uveal melanoma. J. Immunother. Cancer 8, e000341 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raychaudhuri P., Park H. J., FoxM1: A master regulator of tumor metastasis. Cancer Res. 71, 4329–4333 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laissue P., The forkhead-box family of transcription factors: Key molecular players in colorectal cancer pathogenesis. Mol. Cancer 18, 5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen G., Zheng F., Ren D., Du F., Dong Q., Wang Z., Zhao F., Ahmad R., Zhao J., Anlotinib: A novel multi-targeting tyrosine kinase inhibitor in clinical development. J. Hematol. Oncol. 11, 120 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Driehuis E., Kolders S., Spelier S., Lõhmussaar K., Willems S. M., Devriese L. A., de Bree R., de Ruiter E. J., Korving J., Begthel H., van Es J. H., Geurts V., He G.-W., van Jaarsveld R. H., Oka R., Muraro M. J., Vivié J., Zandvliet M. M. J. M., Hendrickx A. P. A., Iakobachvili N., Sridevi P., Kranenburg O., van Boxtel R., Kops G. J. P. L., Tuveson D. A., Peters P. J., van Oudenaarden A., Clevers H., Oral mucosal organoids as a potential platform for personalized cancer therapy. Cancer Discov. 9, 852–871 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Rambow F., Rogiers A., Marin-Bejar O., Aibar S., Femel J., Dewaele M., Karras P., Brown D., Chang Y. H., Debiec-Rychter M., Adriaens C., Radaelli E., Wolter P., Bechter O., Dummer R., Levesque M., Piris A., Frederick D. T., Boland G., Flaherty K. T., van den Oord J., Voet T., Aerts S., Lund A. W., Marine J.-C., Toward minimal residual disease-directed therapy in melanoma. Cell 174, 843–855.e19 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Tsoi J., Robert L., Paraiso K., Galvan C., Sheu K. M., Lay J., Wong D. J. L., Atefi M., Shirazi R., Wang X., Braas D., Grasso C. S., Palaskas N., Ribas A., Graeber T. G., Multi-stage differentiation defines melanoma subtypes with differential vulnerability to drug-induced iron-dependent oxidative stress. Cancer Cell 33, 890–904.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dudley M. E., Wunderlich J. R., Shelton T. E., Even J., Rosenberg S. A., Generation of tumor-infiltrating lymphocyte cultures for use in adoptive transfer therapy for melanoma patients. J. Immunother. 26, 332–342 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arozarena I., Wellbrock C., Phenotype plasticity as enabler of melanoma progression and therapy resistance. Nat. Rev. Cancer 19, 377–391 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Kao H.-F., Liao B.-C., Huang Y.-L., Huang H.-C., Chen C.-N., Chen T.-C., Hong Y.-J., Chan C.-Y., Chia J.-S., Hong R.-L., Afatinib and pembrolizumab for recurrent or metastatic head and neck squamous cell carcinoma (ALPHA Study): A Phase II study with biomarker analysis. Clin. Cancer Res. 28, 1560–1571 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalaora S., Nagler A., Wargo J. A., Samuels Y., Mechanisms of immune activation and regulation: Lessons from melanoma. Nat. Rev. Cancer 22, 195–207 (2022). [DOI] [PubMed] [Google Scholar]
- 43.Newell F., da Silva I. P., Johansson P. A., Menzies A. M., Wilmott J. S., Addala V., Carlino M. S., Rizos H., Nones K., Edwards J. J., Lakis V., Kazakoff S. H., Mukhopadhyay P., Ferguson P. M., Leonard C., Koufariotis L. T., Wood S., Blank C. U., Thompson J. F., Spillane A. J., Saw R. P. M., Shannon K. F., Pearson J. V., Mann G. J., Hayward N. K., Scolyer R. A., Waddell N., Long G. V., Multiomic profiling of checkpoint inhibitor-treated melanoma: Identifying predictors of response and resistance, and markers of biological discordance. Cancer Cell 40, 88–102.e7 (2022). [DOI] [PubMed] [Google Scholar]
- 44.Huang F., Santinon F., González R. E. F., Del Rincón S. V., Melanoma plasticity: Promoter of metastasis and resistance to therapy. Front Oncol. 11, 756001 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rambow F., Marine J. C., Goding C. R., Melanoma plasticity and phenotypic diversity: Therapeutic barriers and opportunities. Genes Dev. 33, 1295–1318 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wouters J., Kalender-Atak Z., Minnoye L., Spanier K. I., De Waegeneer M., González-Blas C. B., Mauduit D., Davie K., Hulselmans G., Najem A., Dewaele M., Pedri D., Rambow F., Makhzami S., Christiaens V., Ceyssens F., Ghanem G., Marine J.-C., Poovathingal S., Aerts S., Robust gene expression programs underlie recurrent cell states and phenotype switching in melanoma. Nat. Cell Biol. 22, 986–998 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Kim T. K., Vandsemb E. N., Herbst R. S., Chen L., Adaptive immune resistance at the tumour site: Mechanisms and therapeutic opportunities. Nat. Rev. Drug Discov. 21, 529–540 (2022). [DOI] [PubMed] [Google Scholar]
- 48.D’Angelo S. P., Larkin J., Sosman J. A., Lebbé C., Brady B., Neyns B., Schmidt H., Hassel J. C., Hodi F. S., Lorigan P., Savage K. J., Miller W. H. Jr., Mohr P., Marquez-Rodas I., Charles J., Kaatz M., Sznol M., Weber J. S., Shoushtari A. N., Ruisi M., Jiang J., Wolchok J. D., Efficacy and safety of nivolumab alone or in combination with ipilimumab in patients with mucosal melanoma: A pooled analysis. J. Clin. Oncol. 35, 226–235 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lian B., Si L., Chi Z. H., Sheng X. N., Kong Y., Wang X., Tian H., Li K., Mao L. L., Bai X., Tang B. X., Yan X. Q., Li S. M., Zhou L., Dai J., Tang X. W., Ran F. W., Yao S., Guo J., Cui C. L., Toripalimab (anti-PD-1) versus high-dose interferon-α2b as adjuvant therapy in resected mucosal melanoma: A phase II randomized trial. Ann. Oncol. 33, 1061–1070 (2022). [DOI] [PubMed] [Google Scholar]
- 50.Sheng X., Yan X., Chi Z., Si L., Cui C., Tang B., Li S., Mao L., Lian B., Wang X., Bai X., Zhou L., Kong Y., Dai J., Wang K., Tang X., Zhou H., Wu H., Feng H., Yao S., Flaherty K. T., Guo J., Axitinib in combination with toripalimab, a humanized immunoglobulin G4 monoclonal antibody against programmed cell death-1, in patients with metastatic mucosal melanoma: An open-label phase IB Trial. J. Clin. Oncol. 37, 2987–2999 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang B., Mo J., Yan X., Duan R., Chi Z., Cui C., Si L., Kong Y., Mao L., Li S., Zhou L., Lian B., Wang X., Bai X., Xu H., Li C., Dai J., Guo J., Sheng X., Real-world efficacy and safety of axitinib in combination with anti-programmed cell death-1 antibody for advanced mucosal melanoma. Eur. J. Cancer 156, 83–92 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Boshuizen J., Vredevoogd D. W., Krijgsman O., Ligtenberg M. A., Blankenstein S., de Bruijn B., Frederick D. T., Kenski J. C. N., Parren M., Brüggemann M., Madu M. F., Rozeman E. A., Song J.-Y., Horlings H. M., Blank C. U., van Akkooi A. C. J., Flaherty K. T., Boland G. M., Peeper D. S., Reversal of pre-existing NGFR-driven tumor and immune therapy resistance. Nat. Commun. 11, 3946 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang Z., Lim S.-O., Yan M., Hsu J. L., Yao J., Wei Y., Chang S.-S., Yamaguchi H., Lee H.-H., Ke B., Hsu J.-M., Chan L.-C., Hortobagyi G. N., Yang L., Lin C., Yu D., Hung M.-C., TYRO3 induces anti-PD-1/PD-L1 therapy resistance by limiting innate immunity and tumoral ferroptosis. J. Clin. Invest. 131, e139434 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robertson A. G., Shih J., Yau C., Gibb E. A., Oba J., Mungall K. L., Hess J. M., Uzunangelov V., Walter V., Danilova L., Lichtenberg T. M., Kucherlapati M., Kimes P. K., Tang M., Penson A., Babur O., Akbani R., Bristow C. A., Hoadley K. A., Iype L., Chang M. T.; TCGA Research Network, Cherniack A. D., Benz C., Mills G. B., Verhaak R. G. W., Griewank K. G., Felau I., Zenklusen J. C., Gershenwald J. E., Schoenfield L., Lazar A. J., Abdel-Rahman M. H., Roman-Roman S., Stern M.-H., Cebulla C. M., Williams M. D., Jager M. J., Coupland S. E., Esmaeli B., Kandoth C., Woodman S. E., Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell 32, 204–220.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen S., Zhou Y., Chen Y., Gu J., Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liao Y., Smyth G. K., Shi W., FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Pertea M., Pertea G. M., Antonescu C. M., Chang T.-C., Mendell J. T., Salzberg S. L., StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goldman M. J., Craft B., Hastie M., Repěka K., Dade F. M., Kamath A., Banerjee A., Luo Y., Rogers D., Brooks A. N., Zhu J., Haussler D., Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 38, 675–678 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cibulskis K., Lawrence M. S., Carter S. L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E. S., Getz G., Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boeva V., Popova T., Bleakley K., Chiche P., Cappo J., Schleiermacher G., Janoueix-Lerosey I., Delattre O., Barillot E., Control-FREEC: A tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 28, 423–425 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Legends for tables S1 to S7
Tables S1 to S7