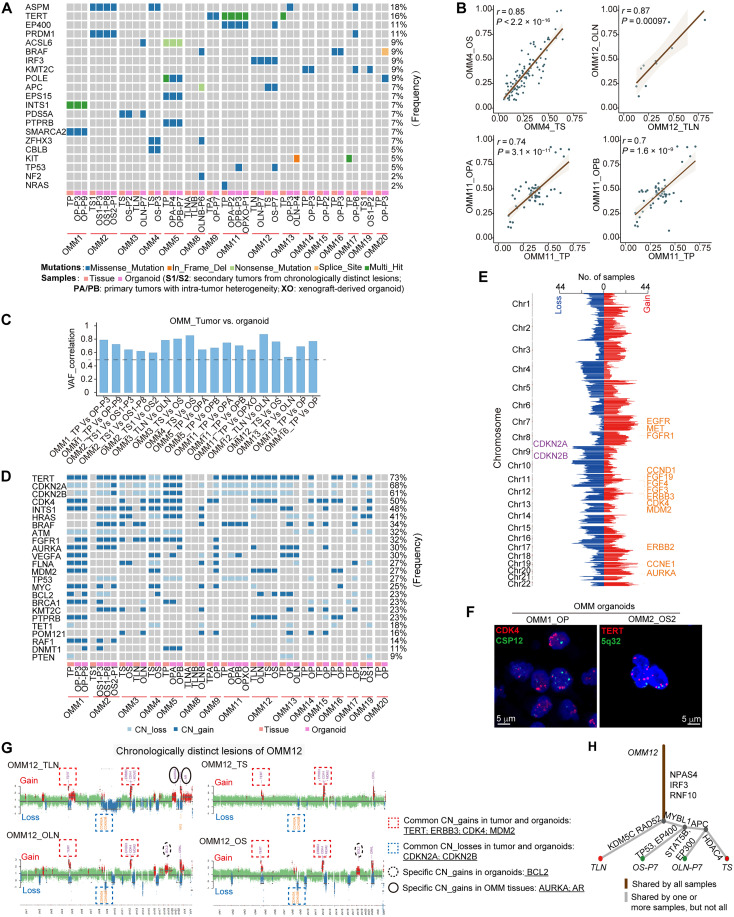
Fig. 2. OMM organoid lines recapitulated major genetic alterations of their parental tumors.
(A) Somatic mutations identified through whole-exon sequencing (WES) in OMM organoids and their parental tumors are summarized. (B) Scatter plots display the correlation between the DNA variant allele frequency (VAF) of OMM organoids and their parental tumors. The figures show the linear regression line (brown) and the corresponding r and P values. (C) Waterfall plots demonstrate the VAF correlation values between OMM organoids and their parental tumors, highlighting significant correlations (P < 0.05). (D) Summary of DNA copy number alterations identified through WES in OMM organoids and their parental tumors. (E) The number of samples with gains (log2Ratio > 0.1) or losses (log2Ratio < −0.1) in corresponding chromosome regions is presented. Chr, chromosome. (F) Representative fluorescence in situ hybridization (FISH) analysis of CDK4 (left) and TERT (right) amplification in OMM organoids. Scale bars, 5 μm. (G) Scatterplots illustrating genome-wide copy number alterations of OMM tissue-organoid pairs in OMM12, with DNA copy number gains (red) and losses (blue) conserved in derived organoid lines. CN_gains, copy number gains; CN_losses, copy number losses. (H) Phylogenetic trees based on somatic mutation variations in OMM12.