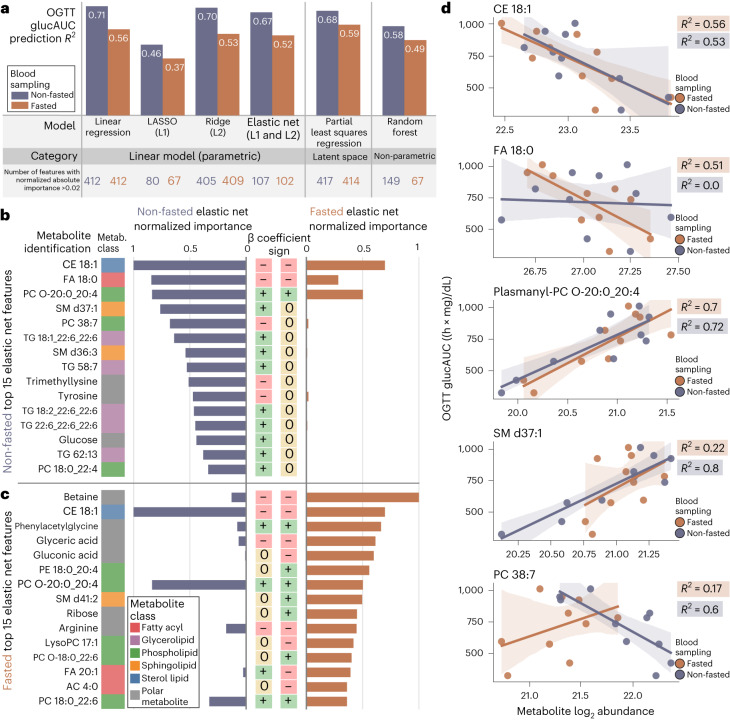
Fig. 4. Linear regression modeling trained on non-fasted plasma samples achieves superior performance in predicting OGTT glucose AUC.
a, Median R2 was compared for six different machine learning model architectures trained on non-fasted and fasted plasma metabolite abundances. Across all models, non-fasted data provided higher median R2 values. Linear regression returned the highest R2, but regularized linear models (LASSO, ridge and elastic net) and other models (PLSr and random forest) were trained to perform feature shrinkage or reduction of feature space dimensionality. Elastic net was most performant for biological interpretation due to its nearly equivalent R2 to linear regression and large coefficient shrinkage. The six models were categorized on the basis of the underlying mechanism of prediction, divided between parametric, latent space and non-parametric methods. b,c, The importance values of the top 15 metabolites in non-fasted (b) and top 15 metabolites in fasted (c) elastic net modeling presented along with importance of molecule in the other sampling method. CE 18:1 and PC O-20:0_20:4 are bolded due to presence in both top 15 lists. d, The top five most important metabolites from non-fasted elastic net modeling were individually regressed to OGTT glucAUC. Dots represent the mean value of each Nile rat’s triplicate metabolite abundance; shaded regions are the 95% bootstrapped confidence interval.