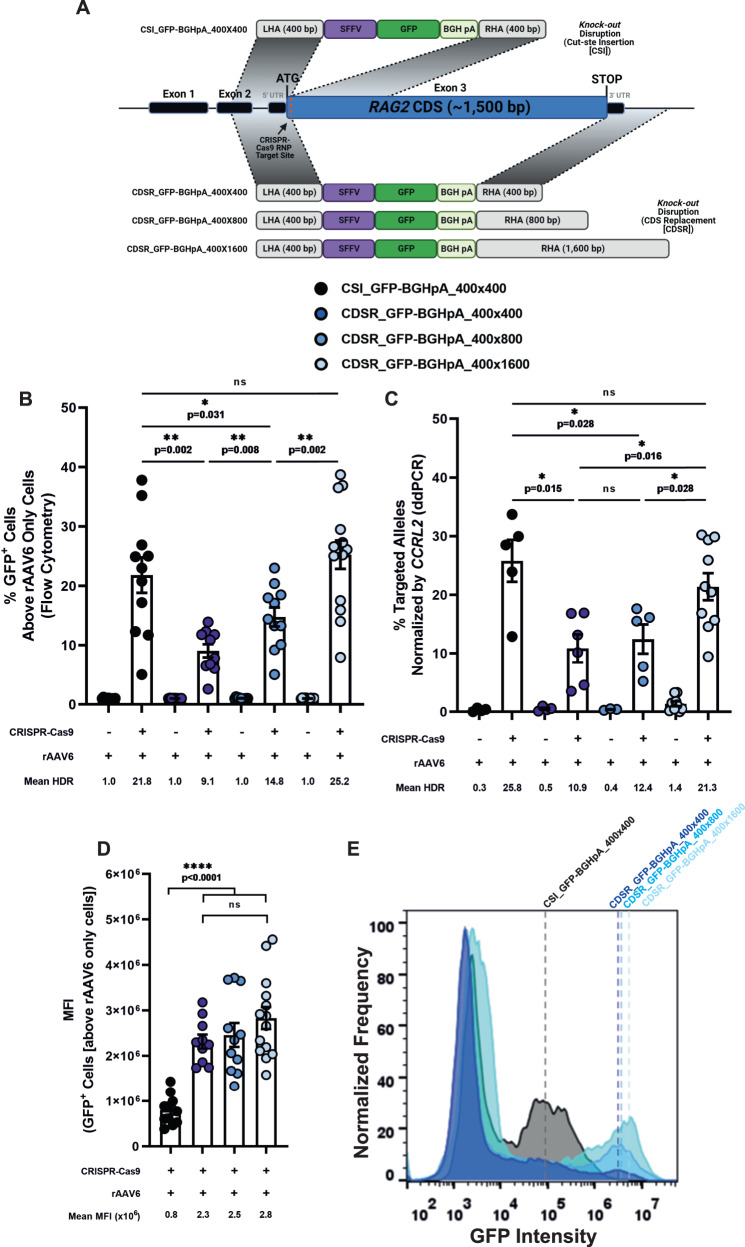
Fig. 1. Different strategies for HDR: cut-site insertion vs. CDS replacement and adjusting homology arms.
A Schematic representation of RAG2 KO disruption donors containing a GFP reporter gene cassette under the control of an SFFV promoter and BGHpA sequence. Successful HDR of the CSI_GFP-BGHpA_400x400 donors results in the integration of the reporter gene approximately 43 bp downstream from the RAG2 ATG start codon. Successful HDR of the three CDSR donors results in the replacement of the entire endogenous RAG2 CDS with the reporter gene cassette. (See Table S1 for a more in-depth description of the donors and Supplementary Data 1 for the exact sequences). Figure was created with BioRender.com. B HDR frequencies analyzed by flow cytometry. See Supplementary Note 1 for a description of the gating strategy. CSI_GFP-BGHpA_400x400 (N = 11), CDSR_GFP-BGHpA_400x400 (N = 10), CDSR_GFP-BGHpA_400x800 (N = 11), CDSR_GFP-BGHpA_400×1600 (N = 14). Data are represented as mean ± SEM. * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001 (Mann-Whitney one-sided test). C Site-specific HDR efficiencies at the RAG2 locus measured by ddPCR and normalized by targeted CCRL2 alleles. CSI_GFP-BGHpA_400×400 ([rAAV6 only: N = 4; CRISPR + AAV: N = 5]), CDSR_GFP-BGHpA_400×400 ([rAAV6 only: N = 4; CRISPR + AAV: N = 6]), CDSR_GFP-BGHpA_400×800 ([rAAV6 only: N = 3; CRISPR + AAV: N = 5]), CDSR_GFP-BGHpA_400×1600 ([rAAV6 only: N = 9; CRISPR + AAV: N = 10]). Data are represented as mean ± SEM. * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001 (Mann-Whitney one-sided test test). D MFI values of HDR+ cells analyzed by flow cytometry. See Supplementary Note 1 for a description of the gating strategy. CSI_GFP-BGHpA_400×400 (N = 11), CDSR_GFP-BGHpA_400×400 (N = 10), CDSR_GFP-BGHpA_400×800 (N = 11), CDSR_GFP-BGHpA_400×1600 (N = 14). Data are represented as mean ± SEM. * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001 (Mann-Whitney one-sided test). Source data are provided as a Source Data file. E Representative histograms of the MFI induced by the four KO donors.