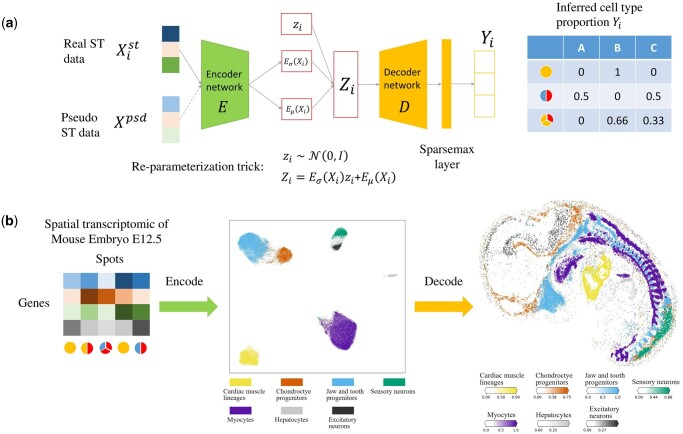
Figure 1.
Overview of stVAE. (a) The network architecture of stVAE. It consists of encoder and decoder networks. stVAE takes gene expression of both pseudo spots and real spatial spots as input and generates their inferred cell type proportions. The pseudo spots are generated from the scRNA-seq reference dataset with known cell type proportions, which can be used as a supervised component to guide the training process for small spatial transcriptomic datasets. (b) An example demonstrating the intuition of stVAE. The encoder network takes spatial transcriptomic spots as inputs and encodes them to the latent features Z. The cell types become better separated at the latent space of Z. The decoder network takes Z as input and estimates the cell type composition of the corresponding spots.