Figure 1. Sampling and analytical approaches used to generate metagenomes, viromes, and vMAGs.
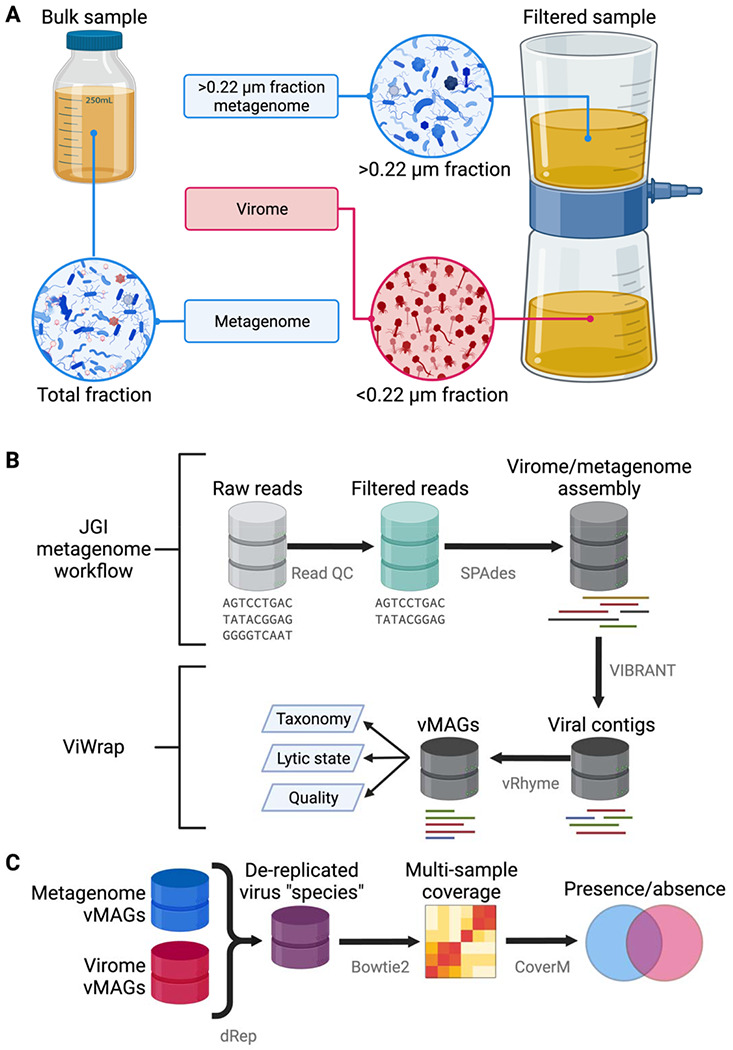
(A) Overview of sampling approaches to generate viromes and metagenomes. Viromes were sequenced from a size fraction below 0.22 μm or from a virus-like particle fraction achieved from ultracentrifugation [11,27]. Metagenomes were sequenced using one of two main approaches: DNA from the bulk sample was extracted and sequenced, allowing the recovery of DNA from prokaryotes, viruses, and other microbes. Alternatively, after filtering a sample to isolate virus-like particles in the <0.22 μm fraction, other studies extracted and sequenced DNA from the remaining >0.22 μm fraction that did not pass through the filter [6,38,39]. (B) Overview of metagenome/virome assembly and virus identification methods to obtain viral metagenome-assembled genomes (vMAGs). (C) Overview of methods for the vMAG presence/absence analysis. Figure created with BioRender.com.
