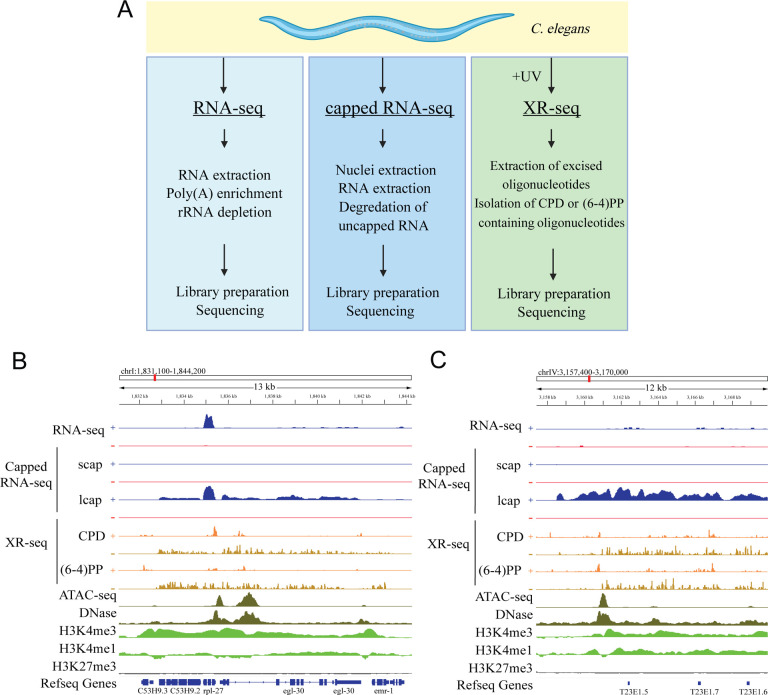
Figure 1. Detection of Transcription-Coupled Repair and Genome-Wide Transcription by XR-seq.
(A) Overview of the study design illustrating the comparative analysis of RNA-seq, capped-RNA-seq, and XR-seq reads for their capacity to identify genome-wide transcription. (B) Distribution of the XR-seq signal over the 13Kb region, separated by strand, for CPD and (6–4)PP 1 hour after 4,000J/m2 UVB treatment. Stranded xpc-1 RNA-seq, long and short capped RNA-seq tracks in blue (plus strand) and red (minus strand) are plotted above, and ATAC-seq (dark green), DNase (dark green), H3K4me3 (light green), H3K4me1 (light green) and H3K27me3 (gray) ChIP-seq tracks are plotted below the XR-seq tracks. Browser view of representative genes clearly demonstrates the occurrence of transcription-coupled repair within the gene body. XR-seq and long-capped RNA-seq methods provide comprehensive coverage of the entire transcript, encompassing both intronic and exonic regions, in annotated genes, in contrast to RNA-seq. The expression of these genes is further substantiated by the presence of high levels of open chromatin and expression-associated markers, including ATAC-seq, DNase-seq, and H3k4me3. The minus strand denotes the transcribed strand, depicted in brown color in the XR-seq representation. (C) Browser view of a representative intergenic region reveals transcription events detected by long-capped RNA-seq and XR-seq but not by RNA-seq. Expression in this intergenic region is corroborated by the presence of elevated levels of open chromatin and expression markers, including ATAC-seq, DNase-seq, H3k4me3, and H3Kme1.