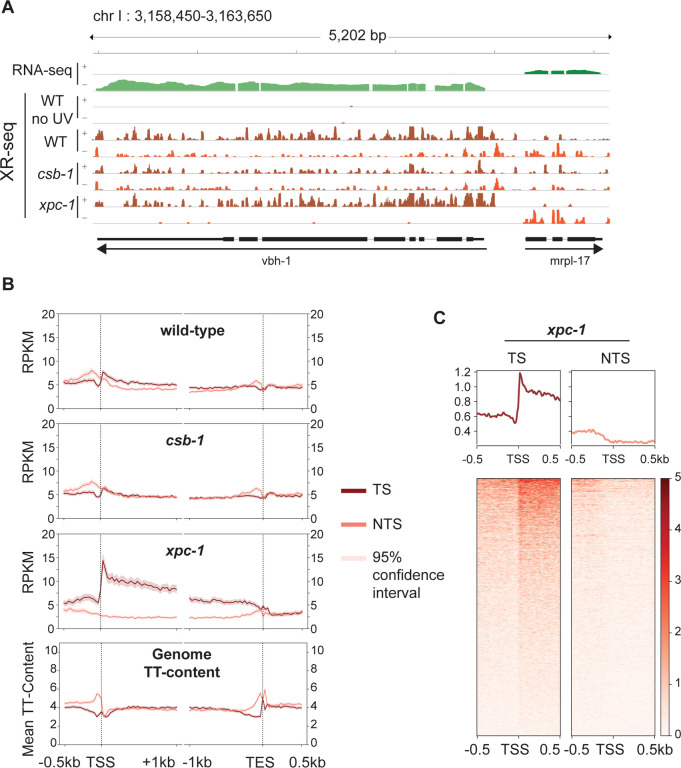
Fig 2. Detection of Transcription-Coupled Repair by XR-seq.
(A) Browser view of the distribution of C. elegans high throughput sequencing reads separated by strand over a representative 5.2 kb region from chromosome I. RNA-seq reads (green) from wild-type (WT) worms is shown on top to illustrate the opposite direction of transcription of the genes vbh-1 and mrpl-17. The strand distribution of XR-seq reads (orange) 1 hour after UV clearly demonstrates the occurrence of transcription-coupled repair within the body of both genes in WT and xpc-1 worms but absent in csb-1. (B) To analyze transcription-coupled repair genome-wide, XR-seq reads on transcribed strand (TS) and non-transcribed strand (NTS) in the indicated strains at 1 hour repair time is plotted with mean reads per kilobase per million mapped reads (RPKM) (y-axis) along the 500 bp upstream and 1 kb downstream of transcription start site (TSS), and 1 kb upstream and 500 bp downstream of transcription end site (TES) (x-axis) for 2,142 genes selected for length > 2 kb and no overlaps with a distance of at least 500 bp between genes. The TT-distribution, as mean TT content (y-axis) was determined for the same gene set and is plotted at the bottom as a measure of expected DNA damage sites. (C) Profile plots and heatmaps of TS and NTS XR-seq reads from the xpc-1 strain at 1 hour repair time spanning the best represented half of 16,588 TSSs > 1 kb apart indicate divergent transcription at promoters.