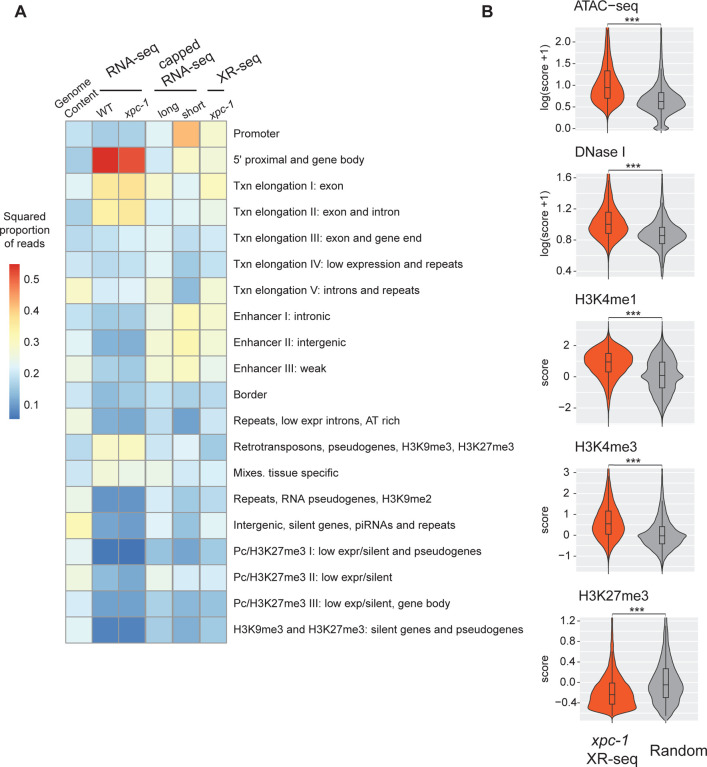
Fig 4. The Transcription-Coupled Repair in Intergenic Regions Detected by xpc-1 XR-seq is Supported by Epigenomic Signatures.
(A) Reads from XR-seq at 1 hour repair time, capped RNA-seq, and RNA-seq were analyzed for overlap with genomic intervals corresponding to 20 distinct predicted chromatin states in C. elegans. The proportion of reads was computed for each of the annotated chromatin states and the square root of the proportion is visualized as a heatmap. (B) Examination of intergenic XR-seq reads, which are undetectable by RNA-seq, in association with ATAC-seq, DNase-seq, H3K4me1, H3K4me3, and H3K27me3 peaks. XR-seq reads exhibit a strong correlation with active transcription markers, in contrast to the repressive marker H4K27me3, when compared to randomly selected genomic regions. All p-values obtained are highly significant (< 2.2e-16) according to nonparametric Wilcoxon rank sum tests.