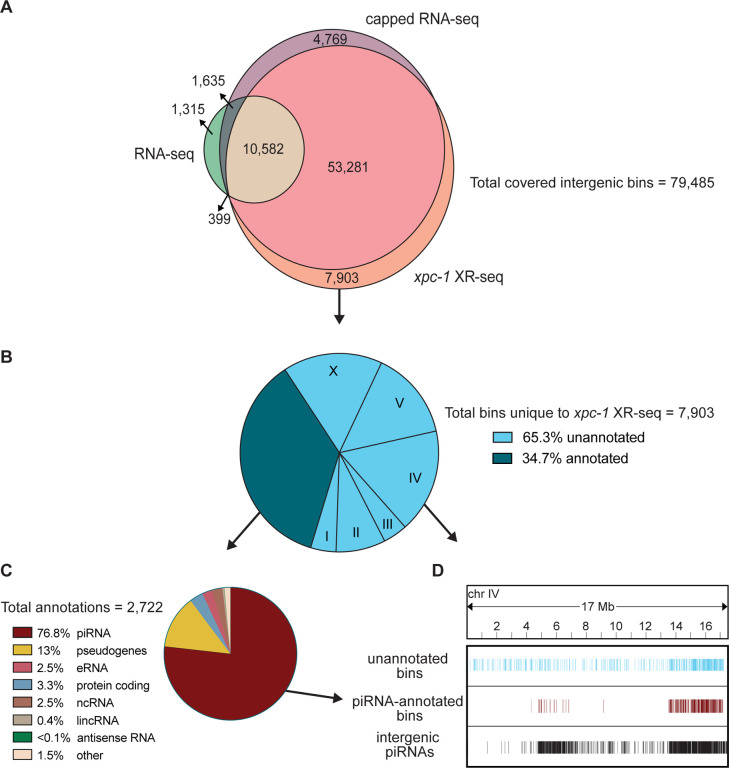
Fig 6. XR-seq identifies intergenic transcription-coupled repair in high concordance with intergenic transcription identified by capped RNA-seq and reveals novel sites of transcription.
For 85,418 intergenic bins, we identified regions with non-zero read counts by short- or long-capped RNA-seq, RNA-seq, and time-course (5mins, 1h, 8h, 16h, 24h, and 48h repair times) xpc-1 XR-seq. (A) Venn diagram of intergenic bins detected by capped RNA-seq, conventional RNA-seq, and xpc-1 XR-seq. To reduce the number of call sets, we required non-zero read counts to be detected: (i) in both replicates for xpc-1 XR-seq; (ii) in both WT and xpc-1 RNA-seq, as they are highly correlated; and (iii) by either short-capped or long-capped RNA-seq, as they are complementary. (B) Pie chart summary of the 7,903 bins unique to xpc-1 XR-seq. 34.7% have been annotated (dark blue) and the remaining 65.3% have not been annotated (light blue) according to the WormBase WS282 annotations. The distribution of chromosomal locations (I-X) is indicated for the unannotated bines. The 68% of annotated bins map to chromosome IV which is not indicated. (C) Pie chart summary of the bins overlapping 2,722 annotations unique to xpc-1 XR-seq dataset. The majority of the unique annotated bins contain piRNAs from chromosome IV, with the remainder consisting of pseudogenes, protein coding regions, eRNAs, lincRNAs, and nRNAs. The ‘other’ category consists of RNAs excluded from the capped RNA-seq dataset (snRNA, tRNAs, rRNAs) only contains 1.5% of bins. (D) Bin distribution along chromosome IV of unique to the xpc-1 XR-seq dataset-unannotated bins (top in blue), unique to the xpc-1 XR-seq dataset-bins with piRNA annotations (middle in burgundy), and intergenic piRNAs from WormBase WS282 annotations (bottom in black).