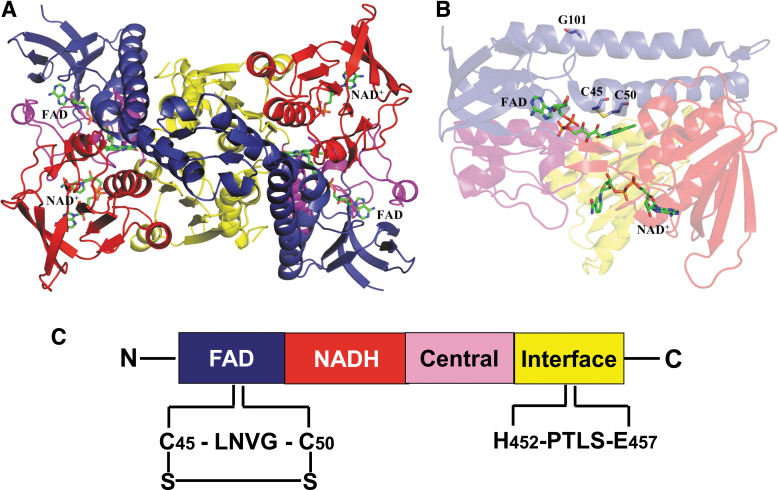
FIG. 3.
A structural model for DLDH. (A) Shows the structure of DLDH homodimer containing a tightly bound FAD molecule and a loosely bound NAD molecule. The two cysteine residues at the active center of the enzyme are shown in (B). FAD-binding domain, the NAD+-binding domain, the central domain, and the interface domain are shown in blue, red, magenta, and yellow, respectively. C45, C50, G101, FAD, and NAD+ are shown as stick model. Images (A, B) were created by Pymol (www.pymol.org) using data from the Protein Data Bank (PDB ID: 1ZMC) (Brautigam et al., 2005). C shows the four domains contained in each monomer from N-terminal to C-terminal and the key amino acid residues involved in NAD-dependent oxidation of dihydrolipoamide. C45, cysteine 45; C50, cysteine 50; FAD, flavin adenine dinucleotide; G101, glycine residue number 101; LNVG, leucine-asparagine-valine-glycine; PTLS, proline-threonine-leucine-serine.