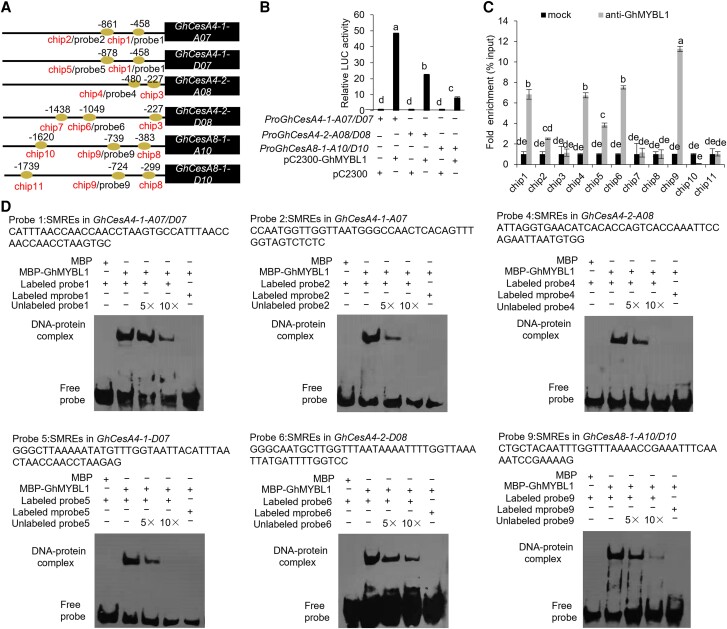
Figure 6.
GhMYBL1 directly binds to the promoters of SCW-related GhCesAs in cotton. A) A schematic diagram depicting SMREs in the promoters of GhCesAs. Horizontal lines represent the promoters, the ovals represent SMREs, the chip1 to chip11 indicate the fragments detected by ChIP-qPCR analysis, and the probe1 to probe9 indicate the fragments detected by EMSA. B) Dual-LUC assay of GhMYBL1 on the promoter activities of GhCesAs (GhCesA4-1-A07/D07, GhCesA4-2-A08/D08, and GhCesA8-1-A10/D10). The relative LUC activities were normalized to the reference REN LUC, and the corresponding effector (+) and empty vector (−) were cofiltrated. The different letters indicate significant difference (P < 0.05) between the 2 groups according to the Tukey HSD test. C) ChIP-qPCR analysis of GhMYBL1's binding to the promoters of GhCesAs. An anti-GhMYBL1 polyclonal antibody was used for ChIP, followed by qPCR analysis of the bound chromatin from 21 DPA cotton fiber cells. The ChIP signal is expressed as the percentage of immunoprecipitated DNA in the total input DNA. Mock, ChIP with control lgG antibody as a negative control. The mean value and Sd were calculated from 3 biological replicates. Values marked with different letters indicate statistically significant differences (P < 0.05) between each group according to the Tukey HSD multiple range test. D) EMSA of GhMYBL1's binding to the SMREs in the promoters of GhCesAs. Biotin-labeled probes were incubated with MBP-GhMYBL1 in vitro. Unlabeled probes were used for competition, and biotin-labeled mutated SMREs were used as negative controls.