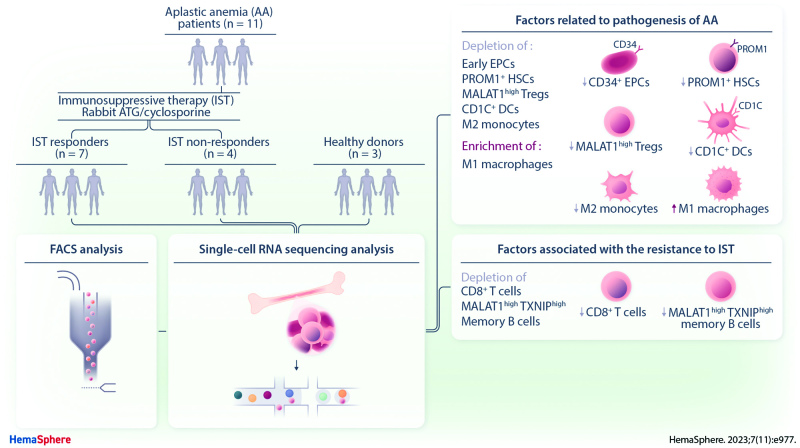
Abstract
Aplastic anemia (AA) is a lethal hematological disorder; however, its pathogenesis is not fully understood. Although immunosuppressive therapy (IST) is a major treatment option for AA, one-third of patients do not respond to IST and its resistance mechanism remains elusive. To understand AA pathogenesis and IST resistance, we performed single-cell RNA sequencing (scRNA-seq) of bone marrow (BM) from healthy controls and patients with AA at diagnosis. We found that CD34+ early-stage erythroid precursor cells and PROM1+ hematopoietic stem cells were significantly depleted in AA, which suggests that the depletion of CD34+ early-stage erythroid precursor cells and PROM1+ hematopoietic stem cells might be one of the major mechanisms for AA pathogenesis related with BM-cell hypoplasia. More importantly, we observed the significant enrichment of CD8+ T cells and T cell–activating intercellular interactions in IST responders, indicating the association between the expansion and activation of T cells and the positive response of IST in AA. Taken together, our findings represent a valuable resource offering novel insights into the cellular heterogeneity in the BM of AA and reveal potential biomarkers for IST, building the foundation for future precision therapies in AA.
INTRODUCTION
Aplastic anemia (AA) is a rare and life-threatening disease characterized by pancytopenia and hypoplastic bone marrow (BM).1 Unlike the inherited AA characterized by BM failure in association with one or more somatic abnormalities, most cases of acquired AA are without any identifiable causes (idiopathic). Although the exact pathogenesis of AA is not completely understood, immune-mediated destruction of hematopoietic stem cells (HSCs) following systemic immune dysregulation has emerged as a main mechanism.2 The most relevant evidence for an immune-mediated mechanism is improvement in blood counts following immunosuppressive therapies (IST).1 The treatment for patients with severe AA (SAA), who are ineligible for hematopoietic stem cell transplantation, aims to suppress the cytotoxic T-lymphocyte response with IST using a combination of antithymocyte globulin (ATG) and cyclosporine.3 However, typical response rates to IST are approximately 60%–70%, and one-third of responders eventually relapse,4 suggesting complex cellular factors linked to AA pathogenesis and the IST resistance mechanism.5
As AA patients have diverse genetic, pathogenic, and immunological backgrounds, it is imperative that the comprehensive characterization of cellular factors for IST response is researched thoroughly. Recently, single-cell RNA sequencing (scRNA-seq) has been widely appreciated in relation to its ability to measure rare or heterogeneous cell populations.6 Lundgren et al highlighted the reduction of cytotoxic STAT3-mutated CD8+ T cells by IST.7 Zhu et al reported the inactivation of cellular interactions mediating the destruction of hematopoietic stem and progenitor cells (HSPCs) by cytotoxic T cells after IST.8 The contribution of changes in B cell gene expression and receptor diversity to AA pathogenesis was also indicated by Tonglin et al.9 However, these studies were limited by the number of either cell types or samples, and a comprehensive understanding of AA pathogenesis and the IST response mechanism is still lacking. Hence, this study analyzed cellular diversity and interactions in the BM of healthy controls and AA patients using scRNA-seq. We identify multiple cellular factors associated with AA pathogenesis and IST resistance.
MATERIALS AND METHODS
Patients
We initially collected consecutive BM samples and clinical data from 18 patients with AA and 10 healthy controls between October 2017 and January 2020 from Seoul St. Mary’s Hospital, Seoul, Republic of Korea (Suppl. Table S1). The samples of 2 of the subjects who were diagnosed with nonsevere AA were excluded from scRNA-seq analysis. The quality control indicated that the BM samples of 5 patients with SAA and 7 healthy controls were inadequate for scRNA-seq analysis. The final cohort included 11 SAA patients and 3 healthy controls and was subjected to scRNA-seq. The diagnosis and severity of idiopathic AA were assessed according to previous reports.10,11 Patients received IST consisting of rabbit ATG (Thymoglobulin; Genzyme-Sanofi, Lyon, France) and cyclosporine (n = 7) as previously described12,13 or cyclosporine monotherapy (n = 4). Patients with SAA received the combination of ATG and cyclosporine as the standard treatment. However, some patients with older age (≥65 y) and/or severe comorbidity related to SAA were treated with cyclosporine monotherapy based on the physician’s discretion. A complete response was defined as hemoglobin ≥10 g/dL, absolute neutrophil count ≥1.0 × 109/L, and platelet count ≥100 × 109/L for >8 weeks without transfusion support. A partial response was determined as no longer meeting the criteria for SAA and having transfusion independence for >8 weeks.3,14 An analysis of glycosylphosphatidylinositol-deficient granulocytes was performed by flow cytometry outlined in our previous report.13 Bone marrow samples of healthy controls were collected from healthy donor for allogeneic BM transplantation following written consent. These were obtained during bone marrow harvesting. The protocol was approved by the institutional review board committees of the Seoul St. Mary’s Hospital (IRB number: KC17TESI0426 and KC23SASI0369). The study was conducted in accordance with the Declaration of Helsinki. The scRNA-seq experiment and flow cytometry analysis were carried out using frozen BM, taken from either AA patients at diagnosis before IST or healthy controls.
Single-cell library preparation and sequencing
scRNA-seq libraries were prepared using the Chromium Single-Cell 3′ Reagent Kits (v2) comprising a Single-Cell 3′ Library & Gel Bead Kit v2 (PN-120237), Single-Cell 3′ Chip Kit v2 (PN-120236), and an i7 Multiplex Kit (PN-120262) (10× Genomics, Pleasanton, CA, USA) and following the Single-Cell 3′ Reagent Kits (v2) User Guide (manual part no. CG00052 Rev A). The libraries were run on a HiSeq X Ten System (Illumina, San Diego, CA, USA) as 150-bp paired-end reads with 1 sample per lane. All the remaining procedures, including library construction, were performed according to the manufacturer’s protocol.
Antibodies and flow cytometry
All antibodies were against human antigens. Anti-TFRC-Alexa Fluor 700 (M-A712), anti-CD4-APC-cy7 (RPA-T4), anti-Prom1-FITC (567029), and anti-CD34 were purchased from BD Biosciences (Franklin Lakes, NJ, USA). Anti-CD3-Brilliant Violet421 (OKT3) and anti-CD8a-APC (300912) were purchased from BioLegend (San Diego, CA, USA). For the phenotypic analysis of BM cells, 1 × 105 cells were suspended in 100 μL Hanks’ balanced salt solution (Mediatech) containing 2% fetal bovine serum. For each sample, cells were incubated with 1 μg of each fluorescent-conjugated primary antibody for 15 minutes at room temperature and washed with Hanks’ balanced salt solution/fetal bovine serum. After being washed with chilled binding buffer, the cells were incubated with streptavidin-PE on ice for 10 minutes before they were washed and analyzed. Staining data were collected using a FACSCantoTM II Cytometer (BD Biosciences).
Clustering analysis and gene set enrichment test
Further scRNA-seq analysis was performed using R (version 4.0.2). We used the FindVariableFeatures in the Seurat package (version 3.2.0) to identify highly variable genes and then performed principal component analysis with the top 2000 variable genes based on the default method, “vst.” Clusters were partitioned with FindClusters in the Seurat package, and cells were projected into a two-dimensional space with uniform manifold approximation and projection based on the default method, “matrix.” The parameters used for clustering analysis are listed in Suppl. Table S2. The differentially expressed genes (DEGs) in each cluster were identified using FindMarkers in the Seurat package based on the default test, “wilcox.” All processes were performed with default options in the Seurat package. We also used the SingleR15 method, which is based on the correlation of gene expression between single cells and a homogenous cell population, with the Novershtern Hematopoietic Data database providing 211 human microarray samples annotated to 16 main types and 38 subtypes of cells. To remove batch effect when integrating the scRNA-seq from the other database with our data, we used the RunHarmony in the Harmony package16 (version 0.1.1) with default parameter while considering the origin of data as batch information, Gene set enrichment test was performed with the Database for Annotation, Visualization, and Integrated Discovery website.17,18 The top 5 gene ontology terms, ranked by P values, are described in our results.
Trajectory analysis
We performed single-cell trajectory analysis using the Monocle2 package (version 2.18.0).19 To chronologically sort cells by pseudotime using the orderCells in Monocle2, we selected the top 2000 DEGs identified by the FindAllMarkers in the Seurat package, based on the default test, “wilcox.” To visualize and interpret the results using the plot_cell_trajectory in Monocle2, the dimension was reduced using reduceDimension in Monocle2 based on the DDRTree method. The other parameters were provided with default options.
Cellular interaction analysis
We performed cell–cell interaction analysis using CellPhoneDB,20 which is a public repository of interactions between ligands and receptors. We used the python (version 3.7.0) and cellphonedb method in CellphoneDB python package (2.1.2) based on the default parameters for the analysis, and the count per million normalized single-cell expression data of HSCs, T cells, and B cells from all samples were used as the input. Interactions of receptors and ligands between 2 cell types were predicted based on the expression profiles of receptors of 1 cell type and the expression profiles of the corresponding ligand of another cell type.
Statistical analysis
All statistical tests were performed on directed pairwise comparisons. Unpaired t-tests were performed to analyze differences between 2 groups. No mathematical corrections were made for multiple comparisons. Significance is displayed as *P < 0.05; **P < 0.01; ***P < 0.001.
RESULTS
Patient characteristics
The median age of the patients was 52 years (range 22–67). Out of total, 8 were classified as SAA, whereas the severity of 3 AA patients was very SAA. Seven patients received IST with ATG and cyclosporine, and 4 patients received cyclosporine monotherapy. Five IST responders (R1–4, 7) achieved a complete response at 8.9, 6.1, 2.3, 11.6, and 24.4 months, respectively. Two IST responders (R5–6) achieved a partial response at 14.3 and 4.3 months, respectively. Four patients had no response (N1–4). Eight patients had small paroxysmal nocturnal hemoglobinuria clones in glycosylphosphatidylinositol-deficient granulocytes (R2–4, R6–7, N2–4). All patients did not have a family history of BM failure syndrome as well as pregnancy/virus-related onset. In 2 patients with onset age younger than 40 years (R2 and R3), there was no evidence of Fanconi anemia by assessment of chromosome breakage on peripheral blood lymphocytes. All patients were classified as having idiopathic AA with normal cytogenetics. Next-generation sequencing-based gene panel testing using peripheral blood did not reveal any pathogenic variants representing inherited BM failure syndrome (Suppl. Tables S3 and S4).21,22
scRNA-seq of BMs from healthy controls, IST responders, and IST non-responders characterizes differential proportion of diverse cell types
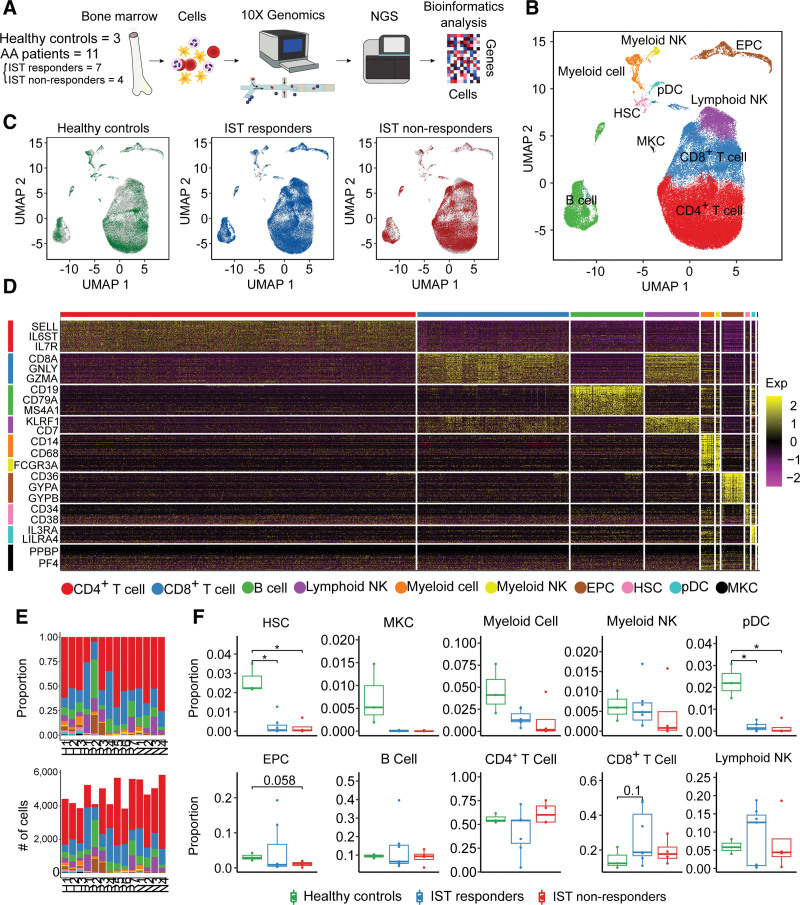
To depict the baseline cellular composition, lineage, and interaction of BM cells from patients with AA and their relation to IST responses, we profiled BM cells from 11 treatment-naive patients with AA, including 7 IST responders (R1–R7), 4 non-responders (N1–N4), and 3 healthy controls (H1–H3) using droplet-based scRNA-seq (Figure 1A). After a quality control, a total of 67,271 cells were retained, comprising 33,646 cells from IST responders (mean: 4807 cells per patient), 21,166 cells from non-responders (mean: 5292 cells per patient), and 12,459 cells from healthy controls (mean: 4153 cells per patient) (Suppl. Table S5). Subsequently, we performed unsupervised clustering analysis and used uniform manifold approximation and projection (UMAP)23 to visualize the cells according to cell type (Figure 1B) and IST response (Figure 1C). We classified our clusters into 8 broad cellular lineages: HSCs, megakaryocytes (MKCs), myeloid cells, myeloid natural killer (NK) cells, plasmacytoid dendritic cells (pDCs), erythroid precursor cells (EPCs), B cells, CD4+ T cells, CD8+ T cells, and lymphoid NK cells by combining the results of the SingleR package in R and the expression profiles of cell type-specific marker genes (Figure 1D). Notably, HSCs and pDCs were significantly depleted in both IST responders and non-responders compared with healthy BM (Figure 1E,F; Suppl. Figure S1; Suppl. Table S6). EPCs were marginally depleted in IST non-responders compared with healthy controls (P = 0.058; Figure 1E,F). We found an increase of CD8+ T cells in IST responders compared with healthy controls (P = 0.1; Figure 1E,F).
Figure 1.
scRNA-seq of healthy controls and patients. (A) Workflow of the study design. scRNA-seq was conducted with bone marrow cells extracted from 3 healthy controls and 11 patients (7 IST responders and 4 non-responders). (B) UMAP plot of clusters including all samples. (C) UMAP plots of the group-specific distribution of cells. (D) Heatmap representing cluster-specific gene expression. The yellow color represents high expression and purple represents low expression. (E) Proportion of clusters (top) and number of cells (bottom) in each sample. (F) Box-and-whisker plot comparing the proportions of clusters among healthy controls, IST responders, and non-responders. The solid horizontal line represents the median, the lower boundary represents the lower quartile, the upper border represents the upper quartile, and the whiskers represent outliers (t test, *P < 0.05). AA = aplastic anemia; EPC = erythroid precursor cell; HSC = hematopoietic stem cell; IST = immunosuppressive therapy; MKC = megakaryocyte; NGS = next generation sequencing; NK = natural killer; pDC = plasmacytoid dendritic cell; scRNA = single-cell RNA; UMAP = uniform manifold approximation and projection.
Depletion of early-stage EPCs was observed in AA patients
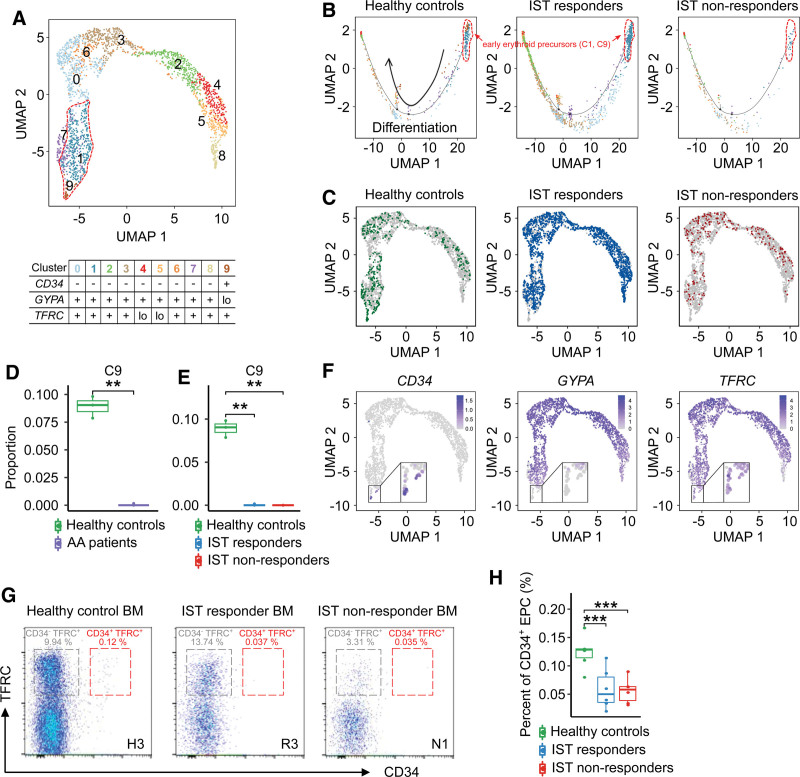
Although the depletion of erythrocytes in the peripheral blood of AA patients was well known,24 the distribution of EPCs in the BM of AA patients is not fully understood. We performed reclustering analysis of EPCs and identified 10 subclusters that were largely grouped based on erythroid genes, such as CD34, GYPA, and TFRC (Figure 2A). Based on our trajectory analysis, they were divided into early-stage (C1, C9), middle-stage (C0, C2, C3, C6, C7, C8), and late-stage (C4, C5) EPCs (Figure 2B; Suppl. Figure S2). Interestingly, the CD34+GYPAloTFRC+ subcluster (C9) was significantly depleted in AA patients (Figure 2C–F; Suppl. Figures S2 and S3). Using flow cytometry, we confirmed the significant depletion of CD34+TFRC+ EPCs in AA patients (Figure 2G,H).
Figure 2.
EPC subclusters. (A) UMAP plot of the EPC subclusters. (B) Trajectory plots of the group-specific distribution of EPC subclusters. The arrow represents the direction of differentiation. (C) UMAP plots of the group-specific distribution of EPCs. (D) Box-and-whisker plot comparing the proportion of cells within EPC subcluster 9 between healthy controls and patients (t-test, ***P < 0.001). (E) Box-and-whisker plot comparing the proportion of cells within EPC subcluster 9 among healthy controls, IST responders, and IST non-responders (t-test, ***P < 0.001). (F) UMAP plots of the expression of marker genes characterizing EPC subclusters (zoomed-in views represent subcluster 9). (G) Representative images for EPC subclusters using flow cytometry. (H) Box-and-whisker plot comparing the percent of CD34+ EPC among healthy controls, IST responders, and non-responders. The solid horizontal line represents the median, the lower boundary represents the lower quartile, the upper border represents the upper quartile, and the whiskers represent outliers (t-test, ***P < 0.001). AA = aplastic anemia; BM = bone marrow; EPC = erythroid precursor cell; IST = immunosuppresive therapy; UMAP = uniform manifold approximation and projection.
HSCs with low expression of PROM1 were partially depleted in AA patients
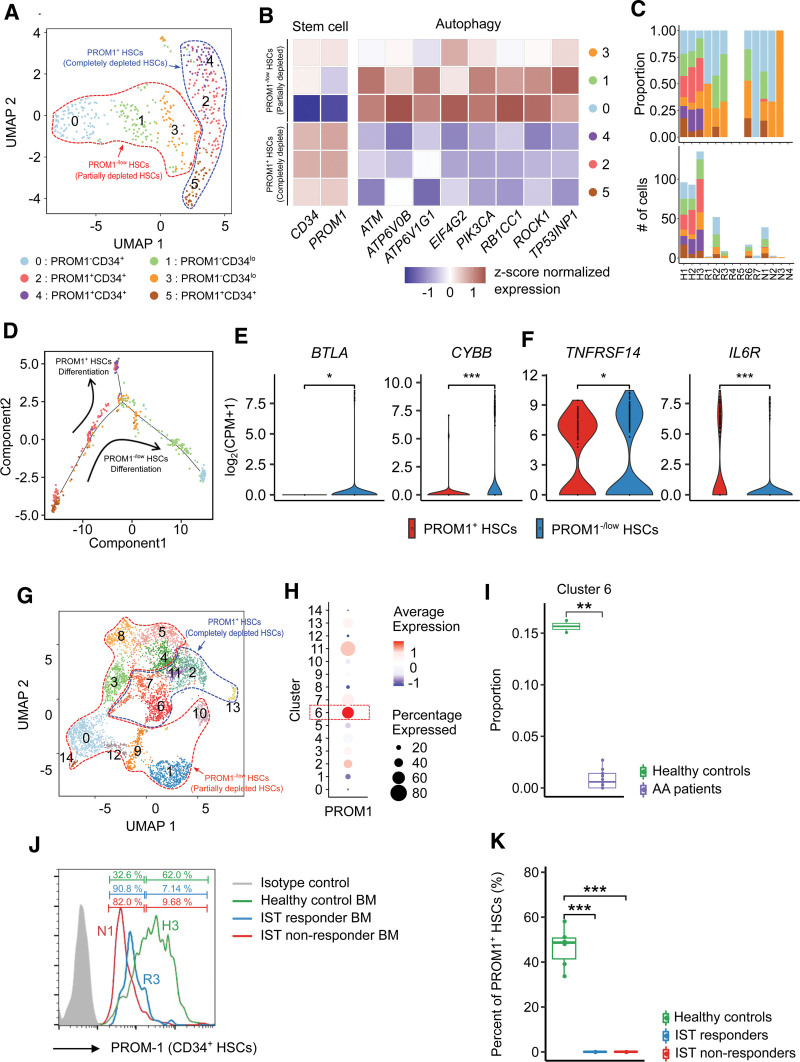
As described above, a significant depletion of HSCs was observed in AA patients. To understand the cellular heterogeneity of HSCs related to this phenomenon, we reclustered HSCs into 6 subclusters, which were largely divided into 2 groups based on PROM1 expression (Figure 3A,B; Suppl. Figure S3).25–27 PROM1+ HSCs (C2, C4, C5) were almost completely depleted in AA patients, but PROM1−/low HSCs (C0, C1, C3) were retained in AA patients (Figure 3B,C). Furthermore, the diversity of HSC subclusters decreased significantly in AA patients compared with healthy controls (Suppl. Figure S4B). Our trajectory analysis showed 2 distinct differentiation pathways for the PROM1+ and PROM1−/low HSC lineages (Figure 3D; Suppl. Figure S5). We analyzed DEGs and found that the partially depleted PROM1−/low HSCs exhibited a relative upregulation of autophagy-related genes implicated in HSC survival (Figure 3B).28 Furthermore, the relative upregulation of inhibitory checkpoints, such as those encoded by BTLA,29,30 CYBB,31,32 and TNFRSF14,33,34 and the downregulation of cytokine receptor IL6R35,36 (Figure 3E) in the partially depleted PROM1−/low HSCs represents the ability of immunosurveillance. To confirm the association of PROM1 to the depletion of HSPCs in AA patients from an independent scRNA-seq cohort, we reclustered HSPCs based on data from Zhu et al (Figure 3G).8 One HSPC subcluster (C6) with relatively high expression of PROM1 was identified as depleted in AA patients (Figure 3H,I), which was consistent with our results. For further validation, we checked the expression of PROM1 in CD34+ hematopoietic cells from healthy controls, IST responders, and non-responders using flow cytometry (Figure 3J). Notably, PROM1 expressing CD34+ hematopoietic cells were significantly depleted in both IST responders and non-responders (Figure 3K).
Figure 3.
HSC subclusters. (A) UMAP plot of HSC subclusters. (B) Heatmap of the expression profile of marker genes [z score of log2(count per million+1)] used to identify the cell type of each subcluster and to determine the degree of depletion. Red and blue colors denote upregulated and downregulated genes, respectively. (C) Proportions (top) and cell counts (bottom) of HSC subclusters for each sample (HSCs were not detected in R4, R5, N3, and N4). (D) Trajectory plot depicting the degree of differentiation and cluster distribution of all samples by pseudotime analysis. The arrow represents the direction of differentiation. (E) Violin plots of the expression profile of DEGs representing the characteristic of PROM1−/low HSCs. (t-test, *P < 0.05, ***P < 0.001). (G) UMAP plot of HSPC subclusters, (H) Dot plot of the expression profile of PROM1. (I) Box-and-whisker plot comparing the proportions of clusters between healthy controls and AA patients. The solid horizontal line represents the median, the lower boundary represents the lower quartile, the upper border represents the upper quartile, and the whiskers represent outliers (t-test, **P < 0.01). (J) Representative histograms show the expression of PROM1 of CD34+ HSCs. (K) Box-and-whisker plot comparing the percent of PROM1+ HSCs among healthy controls, IST responders, and non-responders. The solid horizontal line represents the median, the lower boundary represents the lower quartile, the upper border represents the upper quartile, and the whiskers represent outliers (t-test, ***P < 0.001). AA = aplastic anemia; BM = bone marrow; DEG = differentially expressed genes; HSC = hematopoietic stem cell; HSPC = hematopoietic stem and progenitor cells; IST = immunosuppressive therapy; UMAP = uniform manifold approximation and projection.
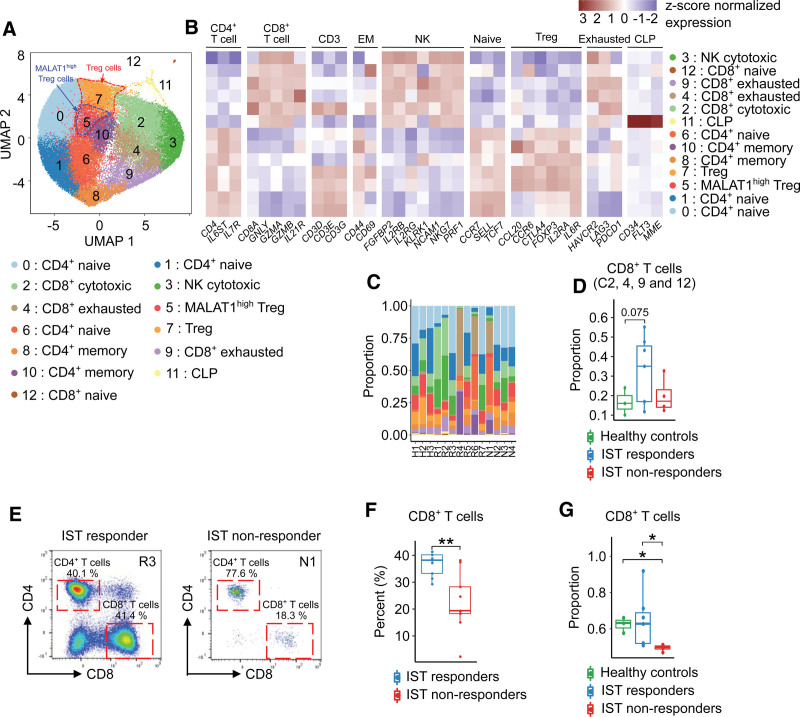
Enrichment of CD8+ T cells in IST responders
We further investigated diverse subclusters of T cells. As a result, we identified 13 T cell subclusters: CD4+ T cells (C0, C1, C6, C8, C10), 4 CD8+ T cells (C2, C4, C9, C12), 1 NK cell (C3), 2 Tregs (C5, C7), and 1 CLP (C11) (Figure 4A) according to the expression patterns of major T cell lineage markers (Figure 4B). Additionally, based on the top markers of each subcluster and the results from the trajectory analysis, we further annotated each subcluster with 4 characteristics, such as naive, cytotoxic, exhausted, and memory (Suppl. Tables S7 and S8 and Figure S6). Although the diversity of the T cell subclusters was similar among healthy controls, IST responders, and non-responders (Suppl. Figure S7A), we found that IST responders showed marginally enriched CD8+ T cells compared with healthy controls (P = 0.075; Figures 1F and 4C,D). Further verification through flow cytometry using 18 BMs of IST responders (n=9) and non-responders (n=9) confirmed that IST responders also showed significantly higher CD8+ T cell rates compared with non-responders (Figure 4E,F). We also found the significant enrichment of CD8+ T cells in IST responders by in silico sorting T cells expressing CD8A from scRNA-seq data (Figure 4G).
Figure 4.
T cell subclusters. (A) UMAP plot of T cell subclusters (NK, Treg, and CLP). (B) Heatmap of the expression of marker genes used to identify the cell type of each subcluster. Red and blue colors denote upregulated and downregulated genes, respectively. (C) Proportion of T cell subclusters in each sample. (D) Box-and-whisker plot comparing the proportion of CD8+ T cell subclusters among healthy controls, IST responders, and non-responders. The solid horizontal line represents the median, the lower boundary represents the lower quartile, the upper border represents the upper quartile, and the whiskers represent outliers (t-test). (E) Representative images of CD4+ T cells and CD8+ T cells from the BMs of IST responders and non-responders through flow cytometry analysis. (F) Box-and-whisker plot comparing the percent of CD8+ T cells between IST responders and IST non-responders. The solid horizontal line represents the median, the lower boundary represents the lower quartile, the upper border represents the upper quartile, and the whiskers represent outliers (t-test, **P < 0.01). (G) Box-and-whisker plot comparing the proportion of CD8+ T cells detected by in silico detection among healthy controls, IST responders, and non-responders. The solid horizontal line represents the median, the lower boundary represents the lower quartile, the upper border represents the upper quartile, and the whiskers represent outliers (t-test, *P < 0.05). BM = bone marrow; CLP = common lymphoid precursor; IST = immunosuppressive therapy; NK = natural killer cell; Treg = regulatory CD4+ T cell; UMAP = uniform manifold approximation and projection.
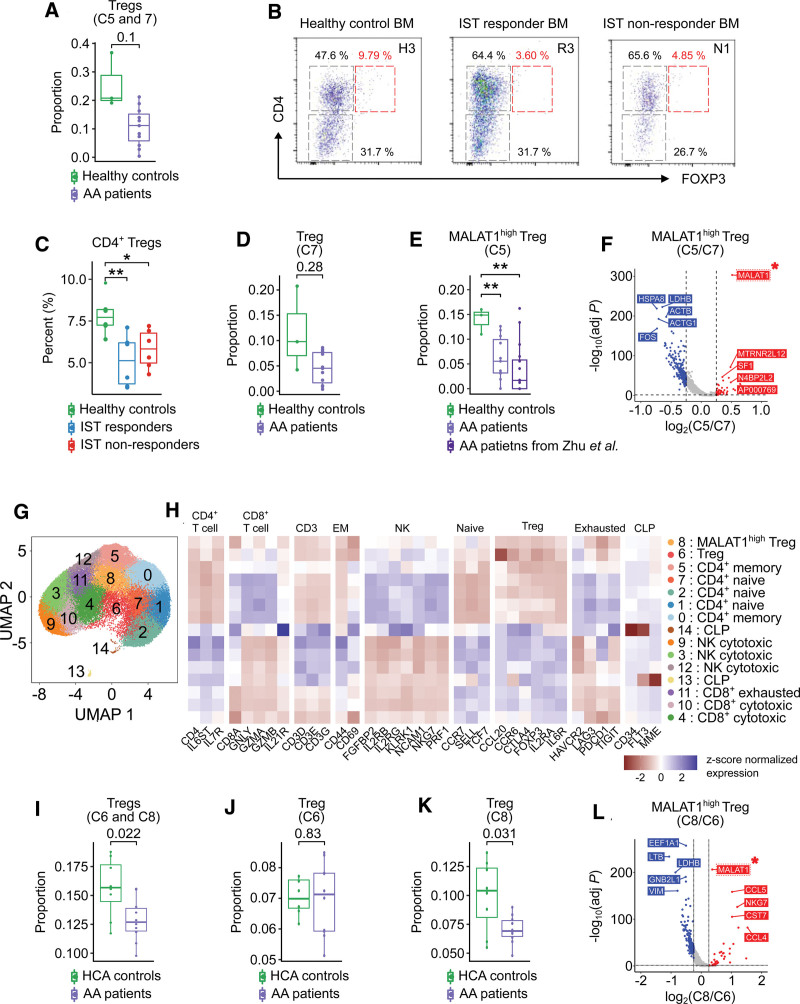
Depletion of MALAT1-upregulated Tregs in AA patients
Next, we investigated the characteristics of Tregs in AA patients and found that Tregs showed the pattern of depletion in AA patients (Figure 5A). The depletion was confirmed by flow cytometry, which measures the proportion of CD4+ Tregs from BM (Figure 5B,C). Importantly, we observed that 1 (C5) of the 2 Treg subclusters was characterized by significant depletion in AA patients compared with healthy controls (Figure 5D,E). To investigate the transcriptomic profile of the C5, we compared DEGs between the 2 Treg subclusters and found that MALAT1 was upregulated in C5 compared with the other Treg subcluster (C7) (Figure 5F; Suppl. Figure S7B). In addition, MALAT1high Tregs showed significant downregulation of FOS and JUNB, which are the genes inducing inflammatory responses in T cells (Figure 5F; Suppl. Figure S7B). Moreover, we verified the depletion of MALAT1high Tregs in AA patients using the independent scRNA-seq data of AA patients obtained from the Zhu et al.8 We identified MALAT1high Treg subcluster from Zhu et al8 based on the Pearson’s coefficient between the expression profiles of the T cells from Zhu et al8 and mean expression profiles of T cell subclusters from our data. The proportion of MALAT1high Treg subcluster from Zhu et al was similar to that of AA patients from our data, representing significant depletion compared with healthy controls from our data (Figure 5E). Additionally, we verified the depletion of MALAT1high Tregs by observing the proportion of MALAT1high Tregs from the integrated scRNA-seq data of healthy BM downloaded from the Human Cell Atlas37 and AA patients from our data after removing batch effect using Harmony.16 We reclustered T cells into 15 subclusters: 5 CD4+ T cell (C0, C1, C2, C5, C7), 2 Treg (C6, C8), 3 CD8+ T cell (C4, C10, C11), 2 NK cell (C3, C9, C12), and 2 common lymphoid precursor (CLP; C13, C14) subclusters (Figure 5G,H). The characteristic of each subcluster was identified based on the results of trajectory analysis (Suppl. Figure S8). We confirmed that the Tregs are significantly depleted in AA patients compared with Human Cell Atlas controls (Figure 5I). Especially, 1 (C8) of 2 Treg subclusters (C6, C8) was characterized with the significant depletion in AA patients and upregulation of MALAT1 compared with the other Treg subcluster (C8) (Figure 5J–F).
Figure 5.
MALAT1high Treg subcluster. (A) Box-and-whisker plot comparing the proportion of Tregs including C5 and C7 between healthy controls and AA patients. The solid horizontal line represents the median, the lower boundary represents the lower quartile, the upper border represents the upper quartile, and the whiskers represent outliers (t-test). (B) Representative images for CD4+ Tregs using flow cytometry. (C) Box-and-whisker plot comparing the percent of CD4+ Tregs among healthy controls, IST responders, and IST non-responders. The solid horizontal line represents the median, the lower boundary represents the lower quartile, the upper border represents the upper quartile, and the whiskers represent outliers (t-test, **P < 0.01 and *P < 0.05). (D) Box-and-whisker plot comparing the proportion of Treg subcluster C7 between healthy controls and AA patients. The solid horizontal line represents the median, the lower boundary represents the lower quartile, the upper border represents the upper quartile, and the whiskers represent outliers (t-test). (E) Box-and-whisker plot comparing the proportion of Treg subcluster C5 among healthy controls and AA patients from our data, and AA patients from Zhu et al. The solid horizontal line represents the median, the lower boundary represents the lower quartile, the upper border represents the upper quartile, and the whiskers represent outliers (t-test, **P < 0.01). (F) Volcano plot for DEGs between T cell subclusters 5 and 7. Red and blue dots denote upregulated and downregulated DEGs, respectively, with an adjusted P value <0.05 and a |log2(FC: fold change)| >0.25. (G) UMAP plot of T cells. (H) Heatmap of the expression of marker genes used to identify the cell type of each subcluster. Red and blue colors denote upregulated and downregulated genes, respectively. (I) UMAP plots displaying the scores associated with immune-suppressive potential of Tregs. (J) Relative expression profiles of MALAT1 among 10 T cell subclusters. (K) Box-and-whisker plot comparing the proportion of cluster 7 of clusters among HCA controls, healthy controls, and AA patients. The solid horizontal line represents the median, the lower boundary represents the lower quartile, the upper border represents the upper quartile, and the whiskers represent outliers (t-test, *P < 0.05). AA = aplastic anemia; BM = bone marrow; DEG = differentially expressed genes; HCA = Human Cell Atlas; IST = immunosuppressive therapy; UMAP = uniform manifold approximation and projection.
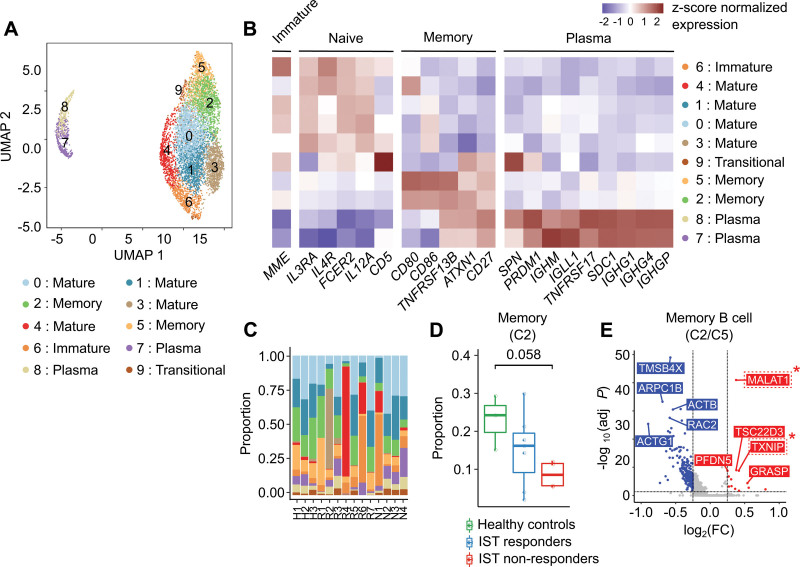
Depletion of activated memory B cells in IST non-responders
After reclustering B cells, 10 subclusters were identified: 1 immature B cell (C6), 4 mature B cells (C0, C1, C3, C4), 2 memory B cells (C2, C5), 1 transitional B cell (C9), and 2 plasma cells (C7, C8) (Figure 6A). The B cell subtypes were labeled according to the expression patterns of major B cell lineage markers (Figure 6B; Suppl. Table S9). The B cell subclusters were largely divided into 2 memory statuses based on the expression of CD27,38 while the expression patterns of IGHG1, IGHG4, and IGLL1 clearly distinguished plasma cells (C6, C9) from the other B cell subclusters (Figure 6A,B; Suppl. Figure S9). Interestingly, 1 memory B cell subcluster (C2) with relative upregulation of MALAT1, which was reported to be associated with viability of diffuse large B cell lymphoma (DLBCL) cell,39 and TXNIP, which is required for expansion of germinal center B cell,40 showed marginally significant depletion in IST non-responders (P = 0.058; Figure 6C–E).
Figure 6.
B cell subclusters. (A) UMAP plot of B cells. (B) Heatmap of the expression of marker genes used to identify the cell type of each subcluster. Red and blue colors denote upregulated and downregulated genes, respectively. (C) Proportion of B cell subclusters in each sample. (D) Box-and-whisker plot comparing the proportion of B cell subcluster 0 among healthy controls, IST responders, and IST non-responders. The solid horizontal line represents the median, the lower boundary represents the lower quartile, the upper border represents the upper quartile, and the whiskers represent outliers (t-test, *P < 0.05). (E) Volcano plot for DEGs between B cell subclusters 2 and 5. Red and blue dots denote upregulated and downregulated DEGs, respectively, with an adjusted P value <0.05 and a |log2(FC: fold change)| >0.25. DEG = differentially expressed genes; IST = immunosuppressive therapy; UMAP = uniform manifold approximation and projection.
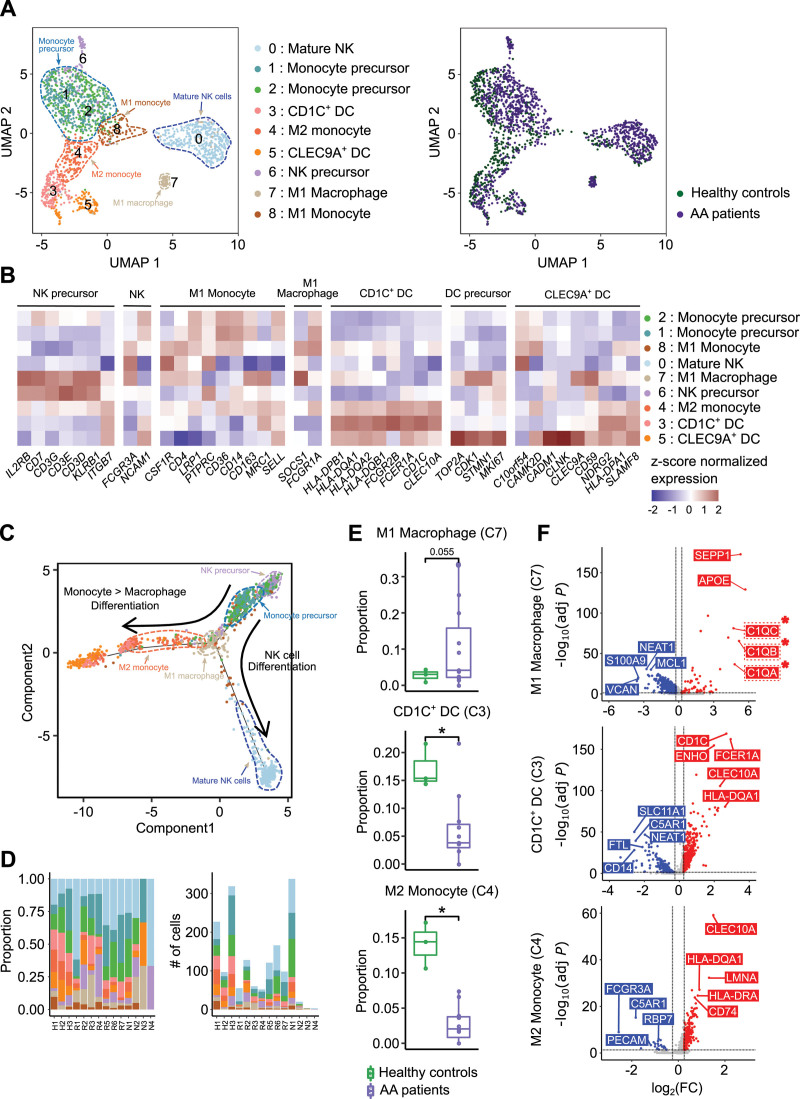
Depletion of CD1C+ DCs and M2 monocytic precursors, and enrichment of inflammatory M1 macrophages in AA patients
When reclustering myeloid cells, we identified 9 subclusters: 1 myeloid NK cells (C0), 1 myeloid NK precursors (C6),41,42 1 M1 macrophage (C7), 1 M1 monocyte (C8), 1 M2 monocyte (C4), 2 monocyte precursors (C1, C2), 1 CD1C+ DC (C3), and 1 CLEC9A+ DC (C5) (Figure 7A–C; Suppl. Table S10).43 Among the 9 subclusters, only 1 subcluster (C7) was marginally increased in AA patients (P = 0.055; Figure 7D,E; Suppl. Figure S10A). C7 was defined as inflammatory M1 macrophage based on the expression of FCGR1A, SOCS1, and complement components (C1QA, C1QB, and C1QC) (Figure 7B,F; Suppl. Figure S10B). In contrast, C3 and C4 were significantly depleted in AA patients and identified as CD1C+ DCs43 and M2 monocytic precursor, respectively, based on the expression profiles of CD1C and CLEC10A (Figure 7B,D–F; Suppl. Figure S10). To further explore the lineages of myeloid cells, we performed trajectory analysis and observed 2 different developmental branches of cellular differentiation (Figure 7C). One branch was from NK precursors (C6) to mature NK cells (C0), and the other was from monocyte precursors (C1, C2) to intermediate-stage M2 monocyte (C4), M1 monocyte (C8), and M1 macrophage (C7) (Figure 7C; Suppl. Figure S11). Late-stage M1 macrophage (C7) was enriched in AA patients, but late-stage CD1C+ DC (C3) and intermediate-stage M2 monocyte (C4) were depleted in AA patients (Figure 7E).
Figure 7.
Myeloid cell subclusters. (A) UMAP plots of all myeloid cell subclusters (NK, DC; left panel) divided by healthy controls and patients (right panel). (B) Heatmap of the expression of marker genes used to identify the cell type of each subcluster. Red and blue colors denote upregulated and downregulated genes, respectively. (C) Trajectory plot describing the degree of differentiation and subcluster distribution by pseudotime analysis. The arrow represents the direction of differentiation. (D) Proportion of myeloid cell subclusters (left) and number of cells (right) in each sample. (E) Box-and-whisker plot comparing the proportion of myeloid cell subclusters 3, 4, and 7 between healthy controls and patients. The solid horizontal line represents the median, the lower boundary represents the lower quartile, the upper border represents the upper quartile, and the whiskers represent outliers (t-test, *P < 0.05). (F) Volcano plots of DEGs of myeloid cell subclusters 3, 4, and 7 compared with the other clusters. Red and blue dots denote upregulated and downregulated DEGs, respectively, with an adjusted P value <0.05 and a |log2(FC: fold change)| >0.25. AA = aplastic anemia; DC = dendritic cell; DEG = differentially expressed genes; NK = natural killer cell; UMAP = uniform manifold approximation and projection.
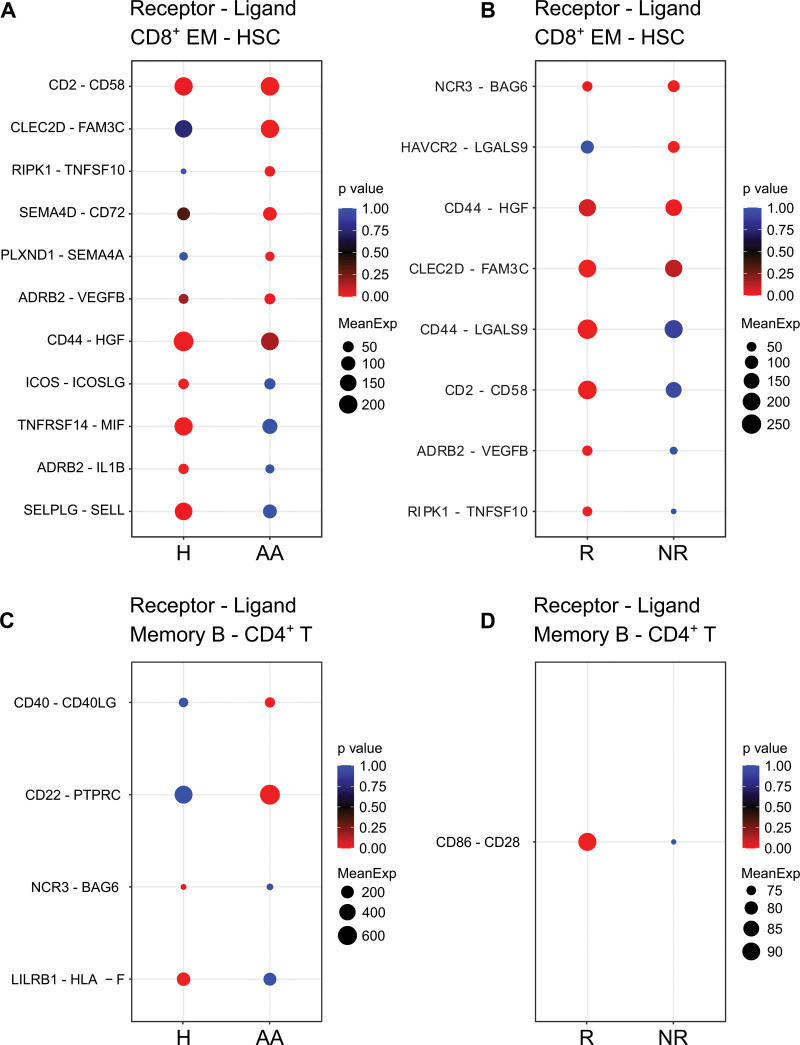
The immune-stimulating intercellular interactions were prominent in IST responders
To gain insight into intercellular interactions associated with AA pathogenesis and IST resistance, we assessed the crosstalk among the major cell types using CellPhoneDB.20 As T cells are major targets of IST and mainly responsible for the depletion of HSCs, our analysis was focused on the intercellular interactions of T cells. We found that several anti-inflammatory interactions between CD8+ EMs and HSCs were inactivated in AA patients. For example, TNFRSF14/MIF34,44 interaction that regulate immune response through the immune checkpoint were inactivated in AA patients (Figure 8A). In contrast, proinflammatory interactions of molecular pairs, such as CLEC2D/FAM3C,45–47 were relatively activated in AA patients (Figure 8A). Additionally, IST responders were characterized by enriched CD2/CD58 interaction, which is essential for cellular immunity48–51 and depleted HAVCR2/LGALS9 interaction which is associated with T cell dysfunction and immune escape (Figure 8B).52–54
Figure 8.
Interactions of CD8+ effector memory T cells and memory B cells with HSCs and CD4+ T cells. (A) Cellular interactions between CD8+ EM T cells and HSCs in healthy controls and AA patients. (B) Cellular interactions between CD8+ EM T cells and HSCs in IST responders and non-responders. (C) Cellular interactions between memory B cells (memory B) and CD4+ T cells (CD4+ T) in healthy controls and AA patients. (D) Cellular interactions between memory B cells and CD4+ T cells (CD4+ T) in IST responders and non-responders. AA = aplastic anemia; CD8+ EM = CD8+ effector memory; HSC = hematopoietic stem cell; IST = immunosuppressive therapy.
Furthermore, several intercellular interactions between CD4+ T cells and memory B cells were found to be potentially associated with AA pathogenesis and IST resistance. For instance, AA patients were characterized by CD4+ T cell–activating interactions, such as CD40/CD40LG55 and CD22/PTPRC.56,57 On the contrary, immune-regulatory interactions, such as LILRB1/HLA-F,58 were inactivated in AA patients (Figure 8C).59 IST responders, in particular, were featured by costimulatory interaction between CD86 and CD28 when compared with non-responders (Figure 8D).60–62
DISCUSSION
In this study, we demonstrated that the hematologic and immunologic abnormalities of the BM of AA patients were caused by the alterations of various types of cells (Figure 1F). Based on the reclustering analysis, the skewed differentiation of several cell types strongly suggests that complex cellular factors are associated with AA pathogenesis and IST resistance.
To disclose the factors affecting hematologic abnormalities, we focused on the depleted subclusters of EPCs and HSCs in AA patients. One early-stage EPC (C9) was completely depleted in AA patients compared with healthy controls (Figure 2D,E). These corroborate the results of a previous study that reported that immature erythroid precursors are sensitive to apoptotic triggering mediated by the activation of the intrinsic and extrinsic apoptotic pathways, which were exacerbated in AA.63 Our results might be related to the significant reduction of mature erythrocytes in the peripheral blood of AA patients.24
In the case of HSCs, we found 2 major groups based on the expression profile of PROM1 (Figure 3A,B). Although PROM1+ HSCs were completely depleted in AA patients, PROM1−/low HSCs were partially depleted in AA patients before exposure to the IST, indicating their recovering roles after IST (Figure 3C). According to the results of the trajectory analysis, each HSC group was predicted to possess a unique trajectory pattern (Figure 3D). Thus, we concluded that the depletion of HSCs was largely dependent on their differentiation based on PROM1 expression (Figure 3D). Importantly, PROM1−/low HSCs were characterized by the increased expression of autophagy-related genes and the inhibitory checkpoints, indicating their implication in HSC survival (Figure 3B,E).28 These distinct transcription patterns of PROM1−/low HSCs could be associated with their residual capacity of regeneration in the abnormal hematologic environment of AA (Figure 3C).
To evaluate immunologic abnormality in AA patients, we researched the cellular status and interactions of diverse immune cell types. By assessing the ratio of several T cell subtypes, we found that CD8+ T cells were expanded in IST responders compared with non-responders. This result confirms the expansion of CD8+ T cells as a strong factor for IST response in AA patients (Figure 1F,4C–G). We also observed that Tregs were depleted in AA patients supporting the immune-reactive status of T cells in this patient group (Figure 5A–C). Especially, MALAT1high Treg subcluster was significantly reduced in AA patients (Figure 5E,F). MALAT1 was previously reported to impact the differentiation of Th1/Th17 cells into Tregs.64 The MALAT1high Treg subcluster was characterized by the low expression of FOS and JUNB inducing inflammatory responses in T cells (Figure 5F).65 Overall, the altered proportions of CD8+ T cells and MALAT1high Tregs suggest that they could be important cellular components involved in IST response and AA pathogenesis.
We also discovered that 1 memory B cell subcluster (C2) characterized by the upregulation of MALAT1 and TXNIP was significantly depleted in IST non-responders compared with healthy controls (Figure 6C–F). In the case of B cell, MALAT1 is known to be highly expressed in DLBCL patients and the knockdown of MALAT1 induced the apoptosis by arresting cell cycle.39 Furthermore, TXNIP was reported to be associated with the expansion of germinal center B cell.40 This result represents the restricted expansion of memory B cell in IST non-responders and potential contribution of the memory B cells in IST response by enhancing the immune microenvironment of responders.
Next, we observed the altered proportion of 3 myeloid cell subclusters, including M1 macrophage (C7), M2 monocyte precursor (C4), and CD1C+ DC (C3) in AA patients (Figure 7D,E). M1 macrophage subcluster (C7) was enriched in AA patients and was characterized by immune-stimulatory traits based on the upregulation of complement components (C1QA, C1QB, and C1QC) (Figure 7F). In contrast to M1 macrophages, M2 monocytes, which were depleted in AA patients, are known as anti-inflammatory cells and play a role in alleviating inflammation in the context of various diseases.66 Interestingly, CD1C+ DC subcluster (C3) was also depleted in AA patients. CD1C+ DCs have varying roles in different contexts. In several previous studies, CD1C+ DCs were reported to be playing an anti-inflammatory role in a specific condition, such as chronic inflammation.43 However, there are also reports suggesting the inflammatory role of CD1C+ DCs in other contexts.67 These data suggest that the enrichment of inflammatory M1 monocytes and the depletion of anti-inflammatory M2 monocytes and CD1C+ DCs are immunological factors of AA pathogenesis.
Apart from myeloid DC cells, we also detected a significant reduction of pDCs in AA patients compared with healthy controls (Figure 1F). Previous studies reported that pDCs are related to anti-inflammatory responses in various sites via both innate and adaptive mechanisms by inducing the generation of Tregs.68,69 Although the inflammatory roles of pDCs in systemic lupus erythematosus and psoriasis have been reported in previous studies,70,71 our findings support their potential roles in inducing Treg generation for the regulation of the cytotoxicity of T cells.
To investigate the association between T cells and HSC depletion in AA patients, we focused on the cellular interactions between CD8+ EMs and HSCs. We identified that CLEC2D/FAM3C,45–47 and PLXND1/SEMA4A72,73 interactions, which are known to be implicated in T cell-mediated immunity and immune sensing of cell death, were enriched in AA patients (Figure 8A). In contrast, several interactions, including ICOS/ICOSLG,74,75 ADRB2/IL1B,76,77 and SELPLG/SELL,78,79 which are associated with the regulation of T cell expansion, infiltration, and autoimmunity, were depleted in AA patients (Figure 8A). Interestingly, we also observed that immune checkpoint signaling-related molecular interactions, such as TNFRSF14/MIF34,44 interaction, were deactivated in AA patients (Figure 8B). We further found that the active interaction between CD2 on CD8+ EMs and CD58 from HSCs was a characteristic of IST responders. CD58 is the costimulatory ligand for CD2 and the CD2/CD58 interactions between cancer cells and T cells are known to be essential for cell lysis.48–51 These results support that the depletion of HSCs in AA patients was induced by the enhanced immune-reactive interactions between CD8+ EM and HSCs, while these intercellular interactions also affected the IST response.
In B cells, we observed that several molecular interactions associated with T cell activation were enriched in AA patients. These interactions including CD40/CD40L55 and CD22/PTPRC57 are known to be implicated in T cell priming56 and maintenance,80 respectively (Figure 8C). Furthermore, the interaction between immunosuppressive receptor LILRB1 on memory B cells and HLA-F from CD4+ T cells was absent in AA patients (Figure 8C).58,59 IST responders in particular were characterized by the enhanced interaction between CD86 and CD28, which enhances T cell survival through the upregulation of Bcl-xL, an antiapoptotic molecule (Figure 8D).61,62
In summary, our findings demonstrate that various cellular factors are related to the immune-stimulating microenvironment in the BM of AA patients, and the IST response is significantly correlated with the level of immune activation. The activated immune environment in the AA patients was induced by the depletion of MALA1high Tregs, CD1C+ DCs, and M2 monocytes, and the enrichment of M1 macrophages resulting in the significant depletion of early CD34+ EPCs. Further immune activation in IST responders compared with non-responders was associated with the expansion of CD8+ T cells, the exacerbated depletion of MALAT1high Tregs, and the enrichment of memory B cells. In addition, PROM1+ HSCs showed distinct transcriptional profiles of immune checkpoints representing the alteration of HSCs in AA patients. These results suggested that diverse cell types, such as EPC, HSC, T cell, myeloid cell, and B cell, should be considered to develop novel therapeutic strategy for treatment of AA patients (Suppl. Table S11). Due to the rarity and hypocellularity of SAA, the small sample size and heterogeneous subjects with different types of IST utilized in our study could be a limitation. Furthermore, since the presented data are mainly based on the computational analyses, additional rigorous validations are required for supporting our data in the future. Although the horse ATG is the standard source of ATG as intensive IST, horse ATG is not available in South America and Asia.11,81 Since the current study included patients who received rabbit ATG, our results may need to be interpreted cautiously for institutions using horse ATG. However, this is the first comprehensive BM single-cell transcriptome analysis of AA with support by flow cytometry that identifies diverse cellular factors related to AA pathogenesis and the IST response. Released library data from the current study could support subsequent studies to identify the change of scRNA-seq based-transcriptome marker in SAA patients, especially in those who were treated with thrombopoietin agonist added to IST as a front-line treatment.82,83
AUTHOR CONTRIBUTIONS
CHL, HK, and SL designed the study. MK and YKM performed experiments. JJ, HJ, SK, TH, DWC and SL analyzed scRNA-seq data. S-SP, H-JK, and JWL collected patient samples and cared patients. SL, CHL, and JWL wrote the article with feedback from JJ, HJ, SK, SS, TH, DWC, MK, YKM, S-SP, and HK.
DATA AVAILABILITY
The raw data are available at https://compbio.unist.ac.kr/UNIST-Aplastic_anemia-KRICT-2019-08.
DISCLOSURES
The authors have no conflicts of interest to disclose.
SOURCE OF FUNDING
This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry (IPET) through Animal Disease Management Technology Development Program from the Ministry of Agriculture, Food and Rural Affairs (MAFRA) of the Korean government (grant no. 318070-3), Intramural Research Fund (KK2031-10) of KRICT, and the Future-Leading Project Research Fund (1.200094.01) of UNIST. This research was also supported by Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (NRF-2018R1A6A1A03025810) and the Ministry of Science and ICT (NRF-2020M3A9B6038849 and NRF-2021R1A2C1094009). The biospecimens and data used for this study were provided by the Biobank of Seoul St. Mary’s Hospital, The Catholic University of Korea, a member of the Korea Biobank Network (KBN4_A01).
Supplementary Material
Footnotes
JJ, HK, S-SP, and MK have contributed equally to this work.
Supplemental digital content is available for this article.
REFERENCES
- 1.Young NS. Aplastic anemia. N Engl J Med. 2018;379:1643–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeng Y, Katsanis E. The complex pathophysiology of acquired aplastic anaemia. Clin Exp Immunol. 2015;180:361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shin SH, Lee SE, Lee JW. Recent advances in treatment of aplastic anemia. Korean J Intern Med. 2014;29:713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L, Liu H. Pathogenesis of aplastic anemia. Hematology. 2019;24:559–566. [DOI] [PubMed] [Google Scholar]
- 5.Shin SH, Lee JW. The optimal immunosuppressive therapy for aplastic anemia. Int J Hematol. 2013;97:564–572. [DOI] [PubMed] [Google Scholar]
- 6.Wu AR, Neff NF, Kalisky T, et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods. 2014;11:41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lundgren S, Keranen MAI, Kankainen M, et al. Somatic mutations in lymphocytes in patients with immune-mediated aplastic anemia. Leukemia. 2021;35:1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu C, Lian Y, Wang C, et al. Single-cell transcriptomics dissects hematopoietic cell destruction and T-cell engagement in aplastic anemia. Blood. 2021;138:23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tonglin H, Yanna Z, Xiaoling Y, et al. Single-cell RNA-seq of bone marrow cells in aplastic anemia. Front Genet. 2022;12:745483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bacigalupo A. How I treat acquired aplastic anemia. Blood. 2017;129:1428–1436. [DOI] [PubMed] [Google Scholar]
- 11.Killick SB, Bown N, Cavenagh J, et al. Guidelines for the diagnosis and management of adult aplastic anaemia. Br J Haematol. 2016;172:187–207. [DOI] [PubMed] [Google Scholar]
- 12.Shin SH, Yoon JH, Yahng SA, et al. The efficacy of rabbit antithymocyte globulin with cyclosporine in comparison to horse antithymocyte globulin as a first-line treatment in adult patients with severe aplastic anemia: a single-center retrospective study. Ann Hematol. 2013;92:817–824. [DOI] [PubMed] [Google Scholar]
- 13.Park SS, Cho SY, Han E, et al. Reactivation and dynamics of cytomegalovirus and Epstein-Barr virus after rabbit antithymocyte globulin and cyclosporine for aplastic anemia. Eur J Haematol. 2019;103:433–441. [DOI] [PubMed] [Google Scholar]
- 14.Scheinberg P, Wu CO, Nunez O, et al. Treatment of severe aplastic anemia with a combination of horse antithymocyte globulin and cyclosporine, with or without sirolimus: a prospective randomized study. Haematologica. 2009;94:348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aran D, Looney AP, Liu L, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20:163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korsunsky I, Millard N, Fan J, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 19.Qiu X, Hill A, Packer J, et al. Single-cell mRNA quantification and differential analysis with Census. Nat Methods. 2017;14:309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R.CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020;15:1484–1506. [DOI] [PubMed] [Google Scholar]
- 21.Chung NG, Kim M. Current insights into inherited bone marrow failure syndromes. Korean J Pediatr. 2014;57:337–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galvez E, Vallespin E, Arias-Salgado EG, et al. Next-generation sequencing in bone marrow failure syndromes and isolated cytopenias: experience of the Spanish network on bone marrow failure syndromes. HemaSphere. 2021;5:e539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diaz-Papkovich A, Anderson-Trocme L, Ben-Eghan C, et al. UMAP reveals cryptic population structure and phenotype heterogeneity in large genomic cohorts. PLoS Genet. 2019;15:e1008432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim-Wanner SZ, Bug G, Steinmann J, et al. Erythrocyte depletion from bone marrow: performance evaluation after 50 clinical-scale depletions with spectra optia BMC. J Transl Med. 2017;15:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z. CD133: a stem cell biomarker and beyond. Exp Hematol Oncol. 2013;2:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miraglia S, Godfrey W, Yin AH, et al. A novel five-transmembrane hematopoietic stem cell antigen: isolation, characterization, and molecular cloning. Blood. 1997;90:5013–5021. [PubMed] [Google Scholar]
- 27.Yin AH, Miraglia S, Zanjani ED, et al. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90:5002–5012. [PubMed] [Google Scholar]
- 28.Huang J, Ge M, Lu S, et al. Impaired autophagy in adult bone marrow CD34+ cells of patients with aplastic anemia: possible pathogenic significance. PLoS One. 2016;11:e0149586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang JA, Lu YB, Wang WD, et al. BTLA-expressing dendritic cells in patients with tuberculosis exhibit reduced production of IL-12/IFN-alpha and increased production of IL-4 and TGF-beta, favoring Th2 and foxp3(+) treg polarization. Front Immunol. 2020;11:518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ning Z, Liu K, Xiong H. Roles of BTLA in immunity and immune disorders. Front Immunol. 2021;12:654960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adane B, Ye H, Khan N, et al. The hematopoietic oxidase NOX2 regulates self-renewal of leukemic stem cells. Cell Rep. 2019;27:238–254.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martner A, Aydin E, Hellstrand K. NOX2 in autoimmunity, tumor growth and metastasis. J Pathol. 2019;247:151–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kotsiou E, Okosun J, Besley C, et al. TNFRSF14 aberrations in follicular lymphoma increase clinically significant allogeneic T-cell responses. Blood. 2016;128:72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aubert N, Brunel S, Olive D, et al. Blockade of HVEM for prostate cancer immunotherapy in humanized mice. Cancers (Basel). 2021;13:3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones BE, Maerz MD, Buckner JH. IL-6: a cytokine at the crossroads of autoimmunity. Curr Opin Immunol. 2018;55:9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka T, Kishimoto T. Targeting interleukin-6: all the way to treat autoimmune and inflammatory diseases. Int J Biol Sci. 2012;8:1227–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Regev A, Teichmann SA, Lander ES, et al. The human cell atlas. Elife. 2017;6:e27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agematsu K, Hokibara S, Nagumo H, et al. CD27: a memory B-cell marker. Immunol Today. 2000;21:204–206. [DOI] [PubMed] [Google Scholar]
- 39.Winkle M, Kluiver JL, Diepstra A, et al. Emerging roles for long noncoding RNAs in B-cell development and malignancy. Crit Rev Oncol Hematol. 2017;120:77–85. [DOI] [PubMed] [Google Scholar]
- 40.Muri J, Thut H, Kopf M. The thioredoxin-1 inhibitor Txnip restrains effector T-cell and germinal center B-cell expansion. Eur J Immunol. 2021;51:115–124. [DOI] [PubMed] [Google Scholar]
- 41.Perez SA, Sotiropoulou PA, Gkika DG, et al. A novel myeloid-like NK cell progenitor in human umbilical cord blood. Blood. 2003;101:3444–3450. [DOI] [PubMed] [Google Scholar]
- 42.Grzywacz B, Kataria N, Kataria N, et al. Natural killer-cell differentiation by myeloid progenitors. Blood. 2011;117:3548–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Villani AC, Satija R, Reynolds G, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356:eaah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Z, Zhang S, Li H, et al. Cellular interaction analysis characterizing immunosuppressive microenvironment functions in MM tumorigenesis from precursor stages. Front Genet. 2022;13:844604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lai JJ, Cruz FM, Rock KL. Immune sensing of cell death through recognition of histone sequences by C-type lectin-receptor-2d causes inflammation and tissue injury. Immunity. 2020;52:123–135.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luck K, Kim DK, Lambourne L, et al. A reference map of the human binary protein interactome. Nature. 2020;580:402–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sirvent S, Vallejo A, Corden E, et al. TNFα signalling in the cutaneous immune network instructs local Th17 allergen-specific inflammatory responses in atopic dermatitis. medRxiv. 2021:2021.10.07.21264714. [Google Scholar]
- 48.Shao T, Shi W, Zheng JY, et al. Costimulatory function of Cd58/Cd2 interaction in adaptive humoral immunity in a zebrafish model. Front Immunol. 2018;9:1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quastel M, Dustin M. The CD58-CD2 axis in cancer immune evasion. Nat Rev Immunol. 2022;22:409. [DOI] [PubMed] [Google Scholar]
- 50.Demetriou P, Abu-Shah E, Valvo S, et al. A dynamic CD2-rich compartment at the outer edge of the immunological synapse boosts and integrates signals. Nat Immunol. 2020;21:1232–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hahn WC, Menu E, Bothwell AL, et al. Overlapping but nonidentical binding sites on CD2 for CD58 and a second ligand CD59. Science. 1992;256:1805–1807. [DOI] [PubMed] [Google Scholar]
- 52.Goncalves Silva I, Yasinska IM, Sakhnevych SS, et al. The Tim-3-galectin-9 secretory pathway is involved in the immune escape of human acute myeloid leukemia cells. EBioMedicine. 2017;22:44–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, Wu K, Tao K, et al. Tim-3/galectin-9 signaling pathway mediates T-cell dysfunction and predicts poor prognosis in patients with hepatitis B virus-associated hepatocellular carcinoma. Hepatology. 2012;56:1342–1351. [DOI] [PubMed] [Google Scholar]
- 54.Yasinska IM, Sakhnevych SS, Pavlova L, et al. The tim-3-galectin-9 pathway and its regulatory mechanisms in human breast cancer. Front Immunol. 2019;10:1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Kooten C, Banchereau J. CD40-CD40 ligand. J Leukoc Biol. 2000;67:2–17. [DOI] [PubMed] [Google Scholar]
- 56.Pasqual G, Chudnovskiy A, Tas JMJ, et al. Monitoring T cell-dendritic cell interactions in vivo by intercellular enzymatic labelling. Nature. 2018;553:496–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sgroi D, Koretzky GA, Stamenkovic I. Regulation of CD45 engagement by the B-cell receptor CD22. Proc Natl Acad Sci U S A. 1995;92:4026–4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lepin EJ, Bastin JM, Allan DS, et al. Functional characterization of HLA-F and binding of HLA-F tetramers to ILT2 and ILT4 receptors. Eur J Immunol. 2000;30:3552–3561. [DOI] [PubMed] [Google Scholar]
- 59.Yu H, Liu H, Zhao Y, et al. Upregulated expression of leukocyte immunoglobulin-like receptor A3 in patients with severe aplastic anemia. Exp Ther Med. 2021;21:346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharp RC, Brown ME, Shapiro MR, et al. The immunoregulatory role of the signal regulatory protein family and CD47 signaling pathway in type 1 diabetes. Front Immunol. 2021;12:739048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fuse S, Zhang W, Usherwood EJ. Control of memory CD8+ T cell differentiation by CD80/CD86-CD28 costimulation and restoration by IL-2 during the recall response. J Immunol. 2008;180:1148–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jeannin P, Magistrelli G, Aubry JP, et al. Soluble CD86 is a costimulatory molecule for human T lymphocytes. Immunity. 2000;13:303–312. [DOI] [PubMed] [Google Scholar]
- 63.Testa U. Apoptotic mechanisms in the control of erythropoiesis. Leukemia. 2004;18:1176–1199. [DOI] [PubMed] [Google Scholar]
- 64.Masoumi F, Ghorbani S, Talebi F, et al. Malat1 long noncoding RNA regulates inflammation and leukocyte differentiation in experimental autoimmune encephalomyelitis. J Neuroimmunol. 2019;328:50–59. [DOI] [PubMed] [Google Scholar]
- 65.Kimura MY, Hayashizaki K, Tokoyoda K, et al. Crucial role for CD69 in allergic inflammatory responses: CD69-Myl9 system in the pathogenesis of airway inflammation. Immunol Rev. 2017;278:87–100. [DOI] [PubMed] [Google Scholar]
- 66.Minogue AM. Role of infiltrating monocytes/macrophages in acute and chronic neuroinflammation: Effects on cognition, learning and affective behaviour. Prog Neuropsychopharmacol Biol Psychiatry. 2017;79(Pt A):15–18. [DOI] [PubMed] [Google Scholar]
- 67.Sanchez-Cerrillo I, Landete P, Aldave B, et al. COVID-19 severity associates with pulmonary redistribution of CD1c+ DC and inflammatory transitional and nonclassical monocytes. J Clin Invest. 2020;130:6290–6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3:133–146. [DOI] [PubMed] [Google Scholar]
- 69.Bekeredjian-Ding I, Greil J, Ammann S, et al. Plasmacytoid dendritic cells: neglected regulators of the immune response to staphylococcus aureus. Front Immunol. 2014;5:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chan VS, Nie YJ, Shen N, et al. Distinct roles of myeloid and plasmacytoid dendritic cells in systemic lupus erythematosus. Autoimmun Rev. 2012;11:890–897. [DOI] [PubMed] [Google Scholar]
- 71.Nestle FO, Conrad C, Tun-Kyi A, et al. Plasmacytoid predendritic cells initiate psoriasis through interferon-alpha production. J Exp Med. 2005;202:135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Toyofuku T, Yabuki M, Kamei J, et al. Semaphorin-4A, an activator for T-cell-mediated immunity, suppresses angiogenesis via Plexin-D1. EMBO J. 2007;26:1373–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang L, Song G, Zheng Y, et al. Expression of SEMAPHORIN 4A and its potential role in rheumatoid arthritis. Arthritis Res Ther. 2015;17:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Iwata R, Hyoung Lee J, Hayashi M, et al. ICOSLG-mediated regulatory T-cell expansion and IL-10 production promote progression of glioblastoma. Neuro Oncol. 2020;22:333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang S, Zhu G, Chapoval AI, et al. Costimulation of T cells by B7-H2, a B7-like molecule that binds ICOS. Blood. 2000;96:2808–2813. [PubMed] [Google Scholar]
- 76.Araujo LP, Maricato JT, Guereschi MG, et al. The sympathetic nervous system mitigates CNS autoimmunity via beta2-adrenergic receptor signaling in immune cells. Cell Rep. 2019;28:3120–3130.e5. [DOI] [PubMed] [Google Scholar]
- 77.Bin W, Aksoy MO, Yang Y, et al. IL-1beta enhances beta2-adrenergic receptor expression in human airway epithelial cells by activating PKC. Am J Physiol Lung Cell Mol Physiol. 2001;280:L675–L679. [DOI] [PubMed] [Google Scholar]
- 78.Tinoco R, Carrette F, Barraza ML, et al. PSGL-1 is an immune checkpoint regulator that promotes T cell exhaustion. Immunity. 2016;44:1190–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Andre P, Spertini O, Guia S, et al. Modification of P-selectin glycoprotein ligand-1 with a natural killer cell-restricted sulfated lactosamine creates an alternate ligand for L-selectin. Proc Natl Acad Sci U S A. 2000;97:3400–3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Al Barashdi MA, Ali A, McMullin MF, et al. Protein tyrosine phosphatase receptor type C (PTPRC or CD45). J Clin Pathol. 2021;74:548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Narita A, Zhu X, Muramatsu H, et al. Prospective randomized trial comparing two doses of rabbit anti-thymocyte globulin in patients with severe aplastic anaemia. Br J Haematol. 2019;187:227–237. [DOI] [PubMed] [Google Scholar]
- 82.Peffault de Latour R, Kulasekararaj A, Iacobelli S, et al. Eltrombopag added to immunosuppression in severe aplastic anemia. N Engl J Med. 2022;386:11–23. [DOI] [PubMed] [Google Scholar]
- 83.Patel BA, Groarke EM, Lotter J, et al. Long-term outcomes in patients with severe aplastic anemia treated with immunosuppression and eltrombopag: a phase 2 study. Blood. 2022;139:34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data are available at https://compbio.unist.ac.kr/UNIST-Aplastic_anemia-KRICT-2019-08.