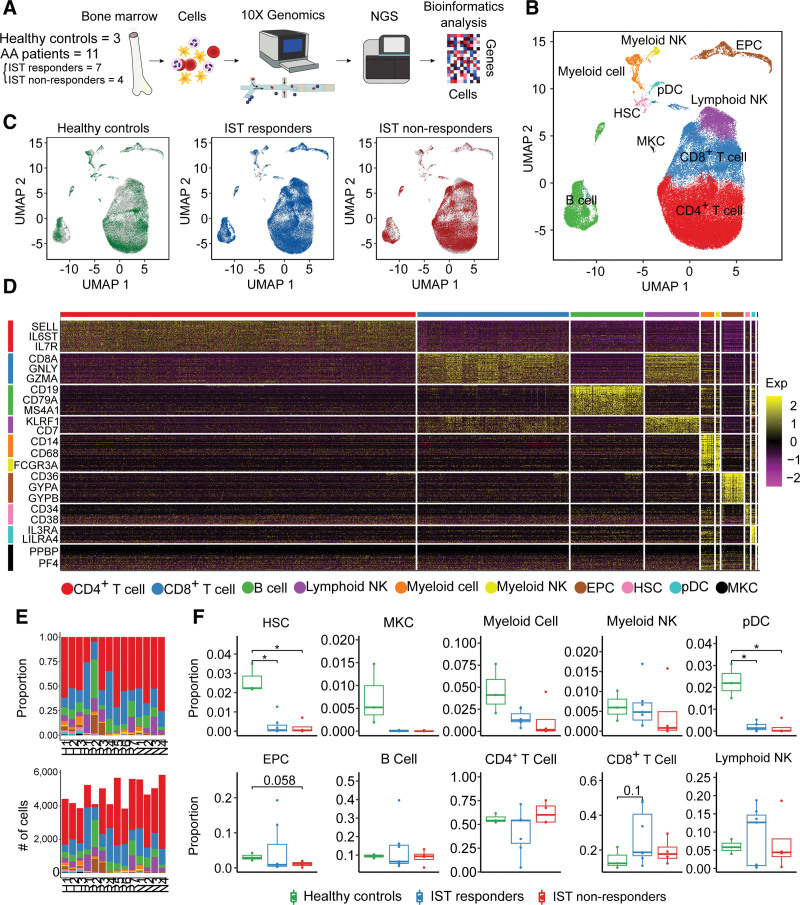
Figure 1.
scRNA-seq of healthy controls and patients. (A) Workflow of the study design. scRNA-seq was conducted with bone marrow cells extracted from 3 healthy controls and 11 patients (7 IST responders and 4 non-responders). (B) UMAP plot of clusters including all samples. (C) UMAP plots of the group-specific distribution of cells. (D) Heatmap representing cluster-specific gene expression. The yellow color represents high expression and purple represents low expression. (E) Proportion of clusters (top) and number of cells (bottom) in each sample. (F) Box-and-whisker plot comparing the proportions of clusters among healthy controls, IST responders, and non-responders. The solid horizontal line represents the median, the lower boundary represents the lower quartile, the upper border represents the upper quartile, and the whiskers represent outliers (t test, *P < 0.05). AA = aplastic anemia; EPC = erythroid precursor cell; HSC = hematopoietic stem cell; IST = immunosuppressive therapy; MKC = megakaryocyte; NGS = next generation sequencing; NK = natural killer; pDC = plasmacytoid dendritic cell; scRNA = single-cell RNA; UMAP = uniform manifold approximation and projection.